✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy Plant Mini Kit (50)
Cat. No. / ID: 74904
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- High-quality total RNA in 30 minutes
- No phenol/chloroform extraction
- No CsCl gradients, no LiCl or ethanol precipitation
- Excellent recovery of RNA
- Ready-to-use RNA for any downstream application
Product Details
The RNeasy Plant Mini Kit includes QIAshredder spin columns for homogenizing and filtering viscous plant or fungal lysates, and RNeasy spin columns for purifying up to 100 μg of high-quality RNA using silica-membrane technology. Purification can be fully automated on the QIAcube Connect. The kit can also be used in combination with a TissueRuptor or TissueLyser system, which provide efficient disruption and homogenization of plant samples.
Performance
The RNeasy Plant Mini Kit is ideal for isolation of total RNA from a wide variety of plant and fungal samples with sample sizes of 10–100 mg tissue, or 100–1 x 107 cells (see table "Selected samples processed with the RNeasy Plant Mini Kit"). The binding capacity of the spin-columns is up to 100 µg RNA. The typical yield from 100 mg plant tissue is between 25 µg and 65 µg of RNA, although amounts of RNA can vary due to the developmental stage and growth conditions of the samples (see table "Yield from 100 mg tissue").
| Plants | Filamentous fungi |
| Anemone sp. Arabidopsis thaliana1 Begonia sp. Beta vulgaris (sugar beet)2 Chlamydomonas reinhardii (unicellular alga) Chrysanthemum Clarkia spp.3 Daucus carota (carrot)4 Diascia sp. Dendranthema sp. Euglena gracilis (unicellular alga) Funaria hygrometrica (moss) Hordeum vulgare (barley) Lycopersicon esculentum (tomato)5 Malus sp. (apple)6 Nicotiana tabacum (tobacco) Oryza sativa (rice) Pelargonium sp. (geranium) Petroselinum crispum (parsley)7 Pisum sativum (pea) Prunus sp. (cherry)6 Ranunculus sp. Ribes nigrum (black currant) Riccia fluitans (liverwort) Sinapis arvensis (mustard) Solanum tuberosum (potato)8 Spinacia oleracea (spinach) Surfinia sp. Triticum aestivum (wheat) Vitis sp. (grapevine)6 Zea mays (maize) |
Acremonium crysogenum9 Fusarium avenaceum9 |
| Sample type | Yield |
| Arabidopsis leaves | 35 µg |
| Maize leaves | 25 µg |
| Tomato leaves | 65 µg |
| Tobacco leaves | 60 µg |
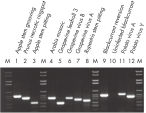
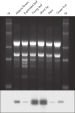
The purified plant RNA, eluted in 30–100 µl volume, includes viral RNA (see figure " Detection of viruses"). All contaminants, such as polysaccharides, are removed, and the eluted RNA is ready for all downstream applications (see figure " Tissue specificity of histone H4 expression").
See figures
Principle
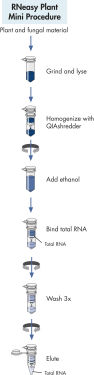
The RNeasy Plant Mini Kit provides QIAshredder columns for homogenization and filtration of viscous plant or fungal lysates by microcentrifugation in combination with RNeasy Mini spin columns for RNA purification. RNeasy technology simplifies total RNA isolation by combining the stringency of guanidine-isothiocyanate lysis with the speed and purity of silica-membrane purification. RNeasy Kits provide the highest-quality RNA with minimum copurification of DNA. For certain RNA applications that are sensitive to very small amounts of DNA, the residual amounts of DNA remaining can be removed using convenient on-column DNase treatment during the RNeasy procedure.
Procedure
See figures
Applications
RNA purified with RNeasy technology is high-quality and is ideal for use in all applications. Downstream applications include:
- Northern, dot, and slot blotting
- End-point RT-PCR
- Quantitative, real-time RT-PCR
- Array analysis
- Poly A+ RNA selection
Supporting data and figures
RNeasy Plant Mini procedure.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, qPCR, real-time RT-PCR, microarray |
| Elution volume | 30–100 µl |
| Sample amount | 10–100 mg |
| Processing | Manual |
| Main sample type | Plant samples |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA |
| Format | Spin column |
| Technology | Silica technology |
| Yield | 25–60 µg |
| Time per run or per prep | 30 minutes |