✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy FFPE Kit (50)
Cat. No. / ID: 73504
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Novel method to overcome formalin crosslinking
- Efficient release of RNA without compromising integrity
- Streamlined protocol providing RNA in just 85 minutes
Product Details
The RNeasy FFPE Kit is specially designed for purifying total RNA from formalin-fixed, paraffin-embedded tissue sections. Special lysis and incubation conditions reverse formaldehye modification of RNA. In addition, the lysis buffer efficiently releases RNA from tissue sections while avoiding further RNA degradation. The kit also uses DNase and DNase Booster Buffer for optimized removal of genomic DNA contamination. RNeasy MinElute spin columns enable purification of total RNA with elution volumes of as low as 10 μl. Purification can be fully automated on the QIAcube Connect.
Performance
See figures
Principle
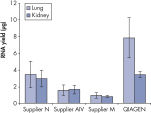
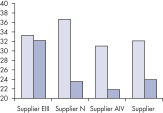
Fixing tissues with formalin leads to RNA–RNA and RNA–protein crosslinking which impairs RNA performance in enzymatic assays. Fixation and embedding conditions can also result in heavily fragmented nucleic acids in FFPE samples. The RNeasy FFPE Kit uses special lysis and incubation conditions to reverse formaldehyde modification of RNA. The lysis buffer efficiently releases RNA from FFPE tissue samples, preventing further RNA degradation. These optimized conditions allow purification of all usable RNA, leading to greater yields from FFPE samples with the RNeasy FFPE Kit than with other methods.
Procedure
The entire RNeasy FFPE procedure can be completed in as little as 85 minutes (see flowchart " RNeasy FFPE procedure"). Sample lysis with proteinase K digestion requires only 15 minutes. After lysis, samples are incubated at 80ºC for 15 minutes. DNase treatment then effectively removes genomic DNA, including small DNA fragments. Finally, concentrated RNA is purified using RNeasy MinElute spin columns, and eluted in a volume of 14–30 µl. Purification can be fully automated on the QIAcube.
See figures
Applications
The RNeasy FFPE Kit isolates all RNA molecules longer than 70 nucleotides from FFPE samples, providing usable RNA fragments for numerous downstream applications, including RT-PCR. However, RNA purified from FFPE samples is heavily fragmented and should not be used in downstream applications that require full-length RNA. Some applications may require modifications to allow the use of fragmented RNA (e.g., designing small amplicons for RT-PCR). For cDNA synthesis, either random or gene-specific primers should be used instead of oligo-dT primers.
Supporting data and figures
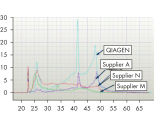
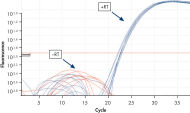
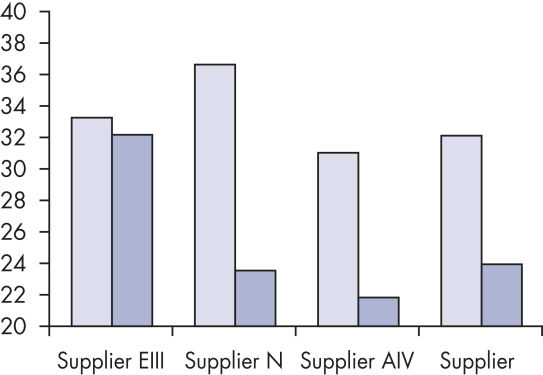
Reliable results in real-time RT-PCR analysis.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, qPCR, real-time RT-PCR, microarray |
| Elution volume | 14–30 µl |
| Main sample type | FFPE tissue samples |
| Processing | Manual (centrifugation), automated (QIAcube) |
| Format | Spin column |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA |
| Time per run or per prep | 70 minutes |
| Sample amount | 1*5 µm to 4*10µm sections |
| Technology | Silica technology |
| Yield | Varies |