HotStarTaq DNA Polymerase (250 U)
Cat. No. / ID: 203203
Features
- Minimal optimization requirements
- High PCR specificity
- Easy handling and room-temperature setup
Product Details
Performance
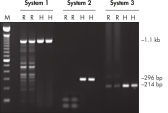
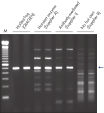
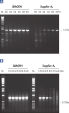
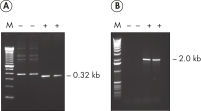
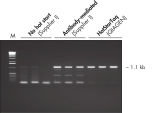
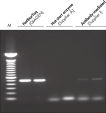
Each lot of HotStarTaq DNA Polymerase is subjected to a comprehensive range of quality control tests, including a stringent PCR specificity and reproducibility assay in which low-copy targets are amplified. HotStarTaq DNA Polymerase outperformed kits tested from other suppliers and ensures high specificity and superior performance in hot-start PCR (see figures " Higher specificity with different primer–template systems" and " Superior performance" and table). The innovative PCR buffer provided with the kit ensures specificity over a wide range of PCR conditions, minimizing the need for optimization (see figure Tolerance to variable temperature and magnesium concentrations). Suboptimal PCR can be improved with Q-Solution, also provided with the kit (see figure " Amplification of difficult templates"). Together, these components ensure specific amplification in a range of applications (see figure " Effect of hot start on RT-PCR performance" and " Highly sensitive single-cell PCR").
| HotStarTaq DNA Polymerase | Hot-start enzyme from Supplier AII | Antibody-mediated | Manual | Wax barrier | |
|---|---|---|---|---|---|
| Specific amplification | ++ | + | + | +/– | +/– |
| Minimal PCR optimization | ++ | +/– | +/– | – | – |
| Easy to use | ++ | ++ | + | – | – |
HotStarTaq DNA Polymerase specifications
Concentration: 5 units/µl
Recombinant enzyme: Yes
Substrate analogs: dNTP, ddNTP, dUTP, biotin-11-dUTP, DIG-11-dUTP, fluorescent-dNTP/ddNTP
Extension rate: 2–4 kb/min at 72°C
Half-life: 10 min at 97°C ; 60 min at 94°C
Amplification efficiency: ≥105 fold
5'–>3' exonuclease activity: Yes
Extra A addition: Yes
3'–>5' exonuclease activity: No
Contaminating nucleases: No
Contaminating RNases: No
Contaminating proteases: No
Self-priming activity: No
See figures
Principle
HotStarTaq DNA Polymerase, a modified form of Taq DNA Polymerase, provides high specificity in hot-start PCR. The kit includes an innovative dual-cation PCR buffer, Q-Solution, and MgCl2.
HotStarTaq DNA Polymerase
HotStarTaq DNA Polymerase is supplied in an inactive state and has no polymerase activity at ambient temperatures. This prevents extension of nonspecifically annealed primers and primer dimers formed at low temperatures during PCR setup and the initial PCR cycle (see figures " Superior performance in hot-start PCR" and " Higher specificity with different primer–template systems"). HotStarTaq DNA Polymerase is activated by a 15-minute incubation at 95°C, which can be incorporated into any existing thermal-cycler program.
QIAGEN PCR Buffer
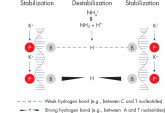
QIAGEN PCR Buffer maintains specific amplification in every cycle of PCR by promoting a high ratio of specific-to-nonspecific primer binding during the annealing step in each PCR cycle (see figure " Increased specificity of primer annealing"). Owing to a uniquely balanced combination of KCl and (NH4)2SO4, the buffer provides stringent primer-annealing conditions over a wider range of annealing temperatures and Mg2+ concentrations than conventional PCR buffers. Optimization of PCR by varying the annealing temperature or the Mg2+ concentration is therefore often minimal or not required (see figure Tolerance to variable temperature and magnesium concentrations).
Q-Solution
Q-Solution, an innovative PCR additive that facilitates amplification of difficult templates by modifying the melting behavior of DNA, is also provided with HotStarTaq DNA Polymerase. This unique reagent improves suboptimal PCR caused by templates that have a high degree of secondary structure or with GC-rich templates (see figure " Amplification of difficult templates"). Unlike other commonly used PCR additives such as DMSO, Q-Solution is used at just one working concentration, is nontoxic, and PCR purity is guaranteed. Adding Q-Solution to the PCR does not compromise PCR fidelity.
See figures
Procedure
See figures
Applications
HotStarTaq DNA Polymerase is suitable for a wide variety of applications, including challenging applications, such as amplification of:
- Complex genomic templates
- Complex cDNA templates (e.g., RT-PCR)
- Very low-copy targets (e.g., single-cell PCR)
- Reactions with multiple primer pairs
Supporting data and figures
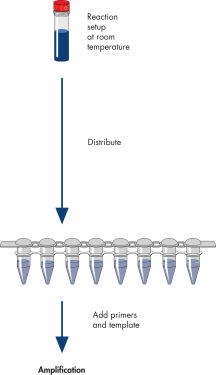
HotStarTaq procedure.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, Complex genomic templates, very low-copy targets |
| With/without hotstart | With hotstart |
| Reaction type | PCR amplification |
| Sample/target type | Genomic DNA and cDNA |
| Real-time or endpoint | Endpoint |
| Enzyme activity | 5' -> 3' exonuclease activity |
| Mastermix | No |
| Single or multiplex | Single |