Configure at GeneGlobe
Find or custom design the right target-specific assays and panels to research your biological targets of interest.
miRCURY LNA miRNA PCR Assay
Cat. No. / ID: 339306
Contains forward and reverse primers for 200 SYBR® Green-based, real-time qPCR reactions, 166 EvaGreen-based digital PCR reactions for Nanoplate 8.5k or 50 EvaGreen-based digital PCR reactions for Nanoplate 26k
Configure at GeneGlobe To see pricing
miRCURY LNA miRNA PCR Assays are intended for molecular biology applications. These products are not intended for the diagnosis, prevention, or treatment of a disease.
Configure at GeneGlobe
Find or custom design the right target-specific assays and panels to research your biological targets of interest.
Features
- Unmatched sensitivity allows miRNA quantification from just 1 pg total RNA
- Great for samples with low RNA yield, such as serum/plasma, FFPE sections and LCM samples
- High specificity discriminates closely related miRNAs and mature miRNA from precursors
- Fast and simple two-step protocol takes less than 3 hours
- Full miRBase coverage enables miRNA profiling from any organism
Product Details
miRCURY LNA miRNA PCR Assays are individual miRNA PCR primer sets that enable extremely sensitive and specific miRNA quantification with the miRCURY LNA miRNA PCR System. Both forward and reverse PCR amplification primers are miRNA-specific and are optimized with LNA technology. Assays are available in tube or plate format with 200 reactions per tube or well.
Need a quote for your research project or would you like to discuss your project with our specialist team? Just contact us!
Performance
Unmatched sensitivity
The exceptional sensitivity of miRCURY LNA miRNA PCR Assays is achieved by combining universal reverse transcription with LNA-enhanced and Tm-normalized primers. This combination enables accurate and reliable quantification of individual miRNAs from as little as 1 pg of total RNA input in the initial first-strand cDNA synthesis (see figure Accurate quantitation from down to 1 pg total RNA starting material).Compared with other miRNA real-time PCR systems that use either stem-loop or standard DNA primers, the LNA-enhanced primers offer significantly increased sensitivity, especially for AT-rich miRNAs (see figure LNA-enhanced primers result in greatly increased sensitivity compared to DNA primers).
The low sample requirements also enable miRNA quantitation using total RNA purified from difficult samples such as LCM samples and serum/plasma (see figures Detection of differential expression of miRNAs in LCM specimens from FFPE tissue sections and Differences in miRNA expression between serum samples).
Fully validated and optimized
All wet-lab validated miRCURY LNA miRNA PCR Assays have been optimized to be as sensitive as possible. Over 80% of assays detect at least 5 miRNA copies in the PCR reaction (see figure Serial dilution of hsa-miR-181a). The primer sets have also been validated for specific amplification of the target and for minimal background signal (see figure Specific and sensitive amplification of miRNA).The only platform with perfect specificity
The miRCURY LNA miRNA PCR Assays are the only miRNA quantification platform with absolute specificity (see the miRQC study published in Nature Methods) and out-performed another probe-based miRNA qPCR platform in specificity tests (see figure Specificity test with probe-based miRNA qPCR system). Perfect specificity eliminates false positives and ensures only robust and reliable miRNA signals.Superior discrimination
The incorporation of LNA in both the forward and reverse PCR amplification primers makes it possible to design assays that can distinguish between miRNA sequences that differ by only one nucleotide (see figure Single-nucleotide discrimination). In addition, the assays can discriminate between mature miRNA sequences and precursor miRNA.Fast, easy and reproducible
The easy-to-follow, 3-hour protocol saves you both time and effort in the laboratory. By using the same RT reaction as the template in all subsequent PCR reactions, the procedure is greatly simplified compared with systems that require miRNA-specific first-strand synthesis. The number of pipetting steps is reduced to a minimum, and technical variation is minimized. This makes it possible to achieve extremely high reproducibility from day-to-day and even site-to-site.miRCURY LNA miRNA PCR Assays have been optimized for use with the miRCURY LNA RT Kit and the miRCURY LNA SYBR Green PCR Kit. Use of other reagents will affect the quality of the results.
See figures
Accurate quantitation from down to 1 pg total RNA starting material. LNA-enhanced primers result in greatly increased sensitivity compared to DNA primers.
LNA-enhanced primers result in greatly increased sensitivity compared to DNA primers. Detection of differential expression of miRNAs in LCM specimens from tissue FFPE sections.
Detection of differential expression of miRNAs in LCM specimens from tissue FFPE sections.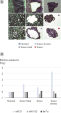 Differences in miRNA expression between serum samples.
Differences in miRNA expression between serum samples.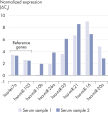 Serial dilution of hsa-miR-181a.
Serial dilution of hsa-miR-181a.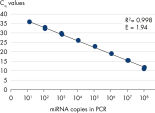 Specific and sensitive amplification of miRNA.
Specific and sensitive amplification of miRNA. Single nucleotide discrimination.
Single nucleotide discrimination.
 LNA-enhanced primers result in greatly increased sensitivity compared to DNA primers.
LNA-enhanced primers result in greatly increased sensitivity compared to DNA primers. Detection of differential expression of miRNAs in LCM specimens from tissue FFPE sections.
Detection of differential expression of miRNAs in LCM specimens from tissue FFPE sections. Differences in miRNA expression between serum samples.
Differences in miRNA expression between serum samples. Serial dilution of hsa-miR-181a.
Serial dilution of hsa-miR-181a. Specific and sensitive amplification of miRNA.
Specific and sensitive amplification of miRNA. Single nucleotide discrimination.
Single nucleotide discrimination.
Principle
A unique system for miRNA profiling
miRCURY LNA miRNA PCR Assays offer the best combination of performance and ease-of-use available on the microRNA real-time PCR market by combining universal RT with LNA PCR amplification (see figure Schematic outline of the miRCURY LNA miRNA PCR System). Universal RT makes it possible to use one first-strand cDNA synthesis reaction as the template for multiple miRNA real-time PCR assays. This saves precious samples, reduces technical variation and saves time in the laboratory. Plus, both the forward and reverse PCR amplification primers are miRNA specific and optimized with LNA. This provides 1) exceptional sensitivity and extremely low background, enabling accurate quantitation of very low miRNA levels and 2) highly specific assays that allow discrimination between closely related miRNA sequences.Coverage
Over 20,000 assays are available covering all organisms in miRBase 20. Over 1,400 assays are fully wet-lab validated for sensitivity, specificity, efficiency and background on both synthetic as well as different biological samples. The remaining assays are in silico-validated using a comprehensive design algorithm that ensures high-quality, species-specific, LNA-enhanced assays with optimal sensitivity and specificity within each organism. This means that several different assays may target the same sequence. Ultimately, the assay for each species is selected based on the genetic background of the organism. If you are working with novel miRNAs, such as from an NGS experiment, custom-designed LNA miRNA primer sets for any miRNA are also available.Reference gene assays for normalization
Nine reference gene primer sets are available, enabling high-quality data normalization and generation of reliable data from many different sample types (see table below). These control primer sets target endogenous, small non-coding RNAs that are constitutively and relatively stably expressed across different human tissues (see figure Expression levels of reference genes in different human tissues). The control reference genes cover a large range of Cq values, offering the possibility to accurately and reliably normalize across a range of miRNA expression levels.The control primer sets have been validated as reference genes for the miRCURY LNA miRNA PCR System and work optimally with the miRCURY LNA RT Kit and the miRCURY LNA SYBR Green PCR Kits. It is very important to always confirm the consistent and unvarying expression of a gene in all tissues and under all treatments of an experiment before choosing it as the reference. We recommend that you test a number of reference genes and use software such as GeNorm and NormFinder (part of the GenEx software) to choose and validate reference genes.
| Product name | Target organism | Alternative names | NCBI reference |
|---|---|---|---|
| Control primer set, SNORD38B (hsa) | hsa | U38B; RNU38B | NR_001457 |
| Control primer set, SNORD44 (hsa) | hsa | U44; RNU44 | NR_002750 |
| Control primer set, SNORD48 (hsa) | hsa | U48; RNU48 | NR_002745 |
| Control primer set, SNORD49A (hsa) | hsa | U49; U49A; RNU49 | NR_002744 |
| Control primer set, SNORA66 (hsa) | hsa | U66; RNU66 | NR_002444 |
| Control primer set, 5S rRNA (hsa) | hsa, mmu | V00589 | |
| Control primer set, U6 snRNA (hsa, mmu) | hsa, mmu, rno | U6 | X59362 |
| Control primer set, RNU5G snRNA (mmu, hsa) | mmu, hsa, rno | Rnu5a; U5a; Rnu5g | NR_002852 |
| Control primer set, RNU1A1 (mmu, hsa) | mmu, hsa, rno | Rnu1a-1; Rnu1a1 | NR_004411 |
| Control primer set, SNORD65 (mmu) | mmu | U65; RNU65 | NR_028541 |
| Control primer set, SNORD68 (mmu) | mmu | U68; RNU68 | NR_028128 |
| Control primer set, SNORD110 (mmu) | mmu | U110; RNU110 | NR_028547 |
See figures
Procedure
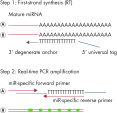
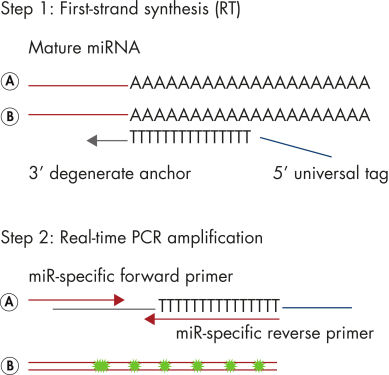
The miRCURY LNA miRNA PCR Assay system is an miRNA-specific, LNA-based system designed for sensitive and accurate detection of miRNA by quantitative real-time PCR using SYBR Green. The first step of the procedure is universal reverse transcription, followed by real-time PCR amplification with LNA-enhanced primers (see figure miRCURY LNA miRNA PCR System at a glance).
Applications
miRCURY LNA miRNA PCR Assays are used as part of the miRCURY LNA miRNA PCR System for:
- Mature miRNA quantification
- snoRNA detection
Supporting data and figures
Schematic outline of the miRCURY LNA miRNA PCR System.
A poly(A) tail is added to the mature miRNA template (step 1A). cDNA is synthesized using a Poly(T) primer with a 3’ degenerate anchor and a 5’ universal tag (step 1B). The cDNA template is then amplified using two miRNA-specific and LNA-enhanced forward and reverse primers (step 2A). SYBR Green is used for detection (step 2B).
Resources
Safety Data Sheets (1)
Quick-Start Protocols (3)
Scientific Posters (1)
Brochures & Guides (3)
Kit Handbooks (3)
Supplementary Protocols (1)
Certificates of Analysis (1)