Features
- Efficient recovery of gDNA from FFPE samples due to optimized lysis
- Enzymatic removal of cytosine deamination artifacts
- Minimized risk of false SNP calls
- Outstanding results in DNA sequencing applications
- Automation on the QIAcube
Product Details
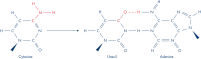
The GeneRead DNA FFPE Kit enables purification of high-quality genomic DNA (gDNA) from formalin-fixed paraffin embedded (FFPE) tissue samples using an optimized silica spin-column–based protocol. Artificial C>T mutations caused by cytosine deamination are critical in next-generation sequencing (NGS). These artifacts are caused by formalin fixation and aging, and result in sequencing errors. The GeneRead FFPE purification procedure also includes enzymatic removal of these artifacts, while ensuring high yields and purity, making it especially suitable for NGS applications.
Purification of DNA using the GeneRead DNA FFPE Kit can be automated on the QIAcube Connect.
Try the next-generation QIAamp DNA FFPE Advanced Kits (Cat.no. 56704 QIAamp DNA FFPE Advanced UNG Kit (50) and Cat. no. 56604 QIAamp FFPE Advanced Kit (50)).
Performance
High yields from limited starting material
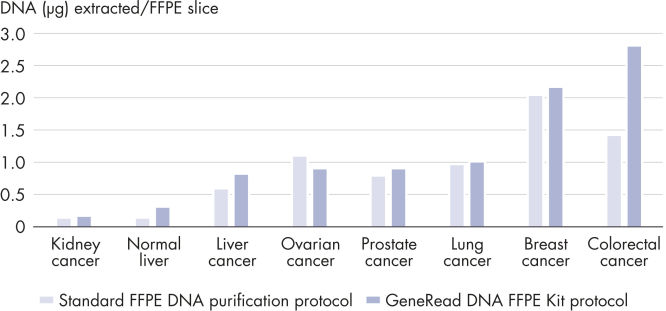
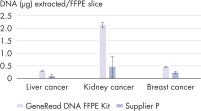
Advances in library preparation and sequencing technologies have made it attractive to perform high-throughput sequencing on large amounts of biobanked FFPE tissues. These technologies have also lowered the frequency threshold at which sequence mutations can be reliably detected. However, sequencing FFPE samples is associated with various challenges. Yields from FFPE samples may be limited due to the compromised status of the DNA. Additionally, these samples are often irreplaceable and there is a need to get the maximum amount of nucleic acid from the smallest amount of starting material. In addition to yield, artifact suppression becomes critical when sequencing FFPE samples, as the relative frequency of false mutations is increased when starting with limited material. The GeneRead DNA FFPE Kit delivers equal or higher yields of double-stranded DNA compared to a standard FFPE DNA isolation protocol (see figure High yields of double-stranded DNA) from small samples (1 x 10 µm slide). The GeneRead DNA FFPE Kit outperforms a kit from another supplier, delivering up to 4-fold higher yields of double-stranded DNA for the samples tested (see figure GeneRead DNA FFPE Kit outperforms a kit from another supplier).
Efficient reduction of artifactual C>T|G>A transitions
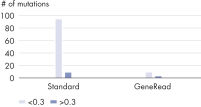
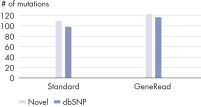
DNA damage caused by formalin fixation and storage is essentially random in nature, with resulting altered sites distributed across the sequence. Only a few copies of the genome will be damaged at any site, leading to a generally low frequency of these artifacts. As low-frequency, novel mutations can be very important, for example in cancer analysis, it is important to distinguish between true and false low-frequency mutations. By examining low-frequency, novel mutations, the number of artifacts in a sample was estimated and the efficacy of the artifact removal process in the GeneRead DNA FFPE protocol was determined. The GeneRead DNA FFPE Kit dramatically reduces low-frequency C>T|G>A transitions while retaining true mutations (see figures Dramatic reduction in artifactual C>T|G>A mutations and Retention of high-frequency C>T|G>A transitions).
Minimizing the risk of false-positive mutations
The removal of artifactual C>T|G>A transitions is especially important when analyzing SNPs associated with cancer. Since deamination mutations occur randomly, they may also be interpreted as cancer-relevant and thereby appear as false-positive SNPs if no artifact removal is employed. The GeneRead DNA FFPE Kit minimizes the risk of false positives as only artifactual mutations are removed (see poster).
See figures
Principle
The GeneRead DNA FFPE Kit provides a streamlined procedure for efficient purification of high yields of DNA from small amounts of FFPE tissue sections. Additionally, the procedure includes the removal of deaminated cytosine to prevent false results in DNA sequencing.
See figures
Procedure
The GeneRead DNA FFPE Kit can also be automated on the QIAcube.
Applications
Supporting data and figures
High yields of double-stranded DNA.