✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy UCP Micro Kit (50)
Cat. No. / ID: 73934
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Ultra-Clean Production (UCP) of spin columns and buffers
- RNA isolation without exogenous nucleic acids for RNA-Seq of low abundance targets
- Consistent RNA yields from very small amounts of starting material
Product Details
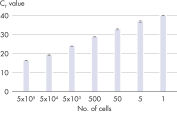
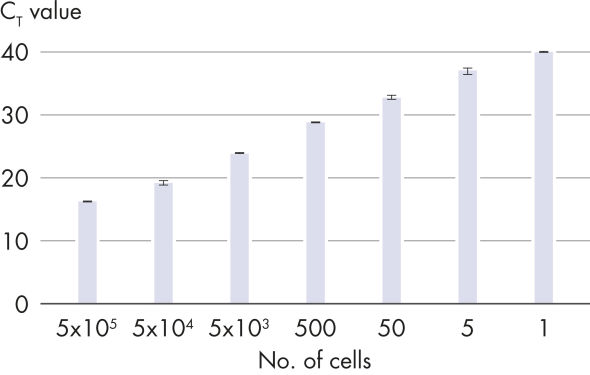
The RNeasy UCP Micro Kit is designed for purification of up to 45 μg RNA from small or low biomass samples. The spin columns and buffers are treated to remove exogenous nucleic acids. RNeasy UCP Micro technology combines the selective binding properties of an ultra-clean, silica-based membrane with the speed of microspin technology. Ultra-clean guanidine-thiocyanate–containing lysis buffer RULT and ethanol are added to the sample to create conditions that promote selective binding of RNA to the RNeasy UCP MinElute membrane. The sample is then applied to the RNeasy UCP MinElute spin column and RNA binds to the silica membrane. Traces of DNA that may copurify are removed by DNase treatment on the RNeasy UCP MinElute spin column. DNase and any contaminants are washed away with ultra-clean wash buffers RUWT and RUPE, and high-quality total RNA is eluted in ultra-clean water (see figures Linearity over a broad dilution range and RNeasy UCP Micro Kit Procedure). With the RNeasy UCP Micro procedure, all RNA molecules longer than 200 nucleotides are purified. The procedure enriches for mRNA, since most RNAs <200 nucleotides are selectively excluded. Tissue samples can be conveniently stabilized using RNAprotect Tissue Reagent or Allprotect Tissue Reagent, and efficiently disrupted using a TissueRuptor or TissueLyser system.
See figures
Performance
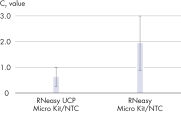
The RNeasy UCP Micro Kit delivers highly reproducible yields of RNA (>200 nt) from small samples. RNA is reliably purified from small numbers of cells, as well as from small amounts of standard tissues. The RNeasy UCP Micro Kit provides maximum sensitivity in quantitative gene expression analyses, such as real-time RT-PCR and NGS, by efficient on-column digestion of genomic DNA. Ultra-clean production minimizes the risk of copurification and subsequent analysis of exogenous nucleic acids (see figures Minimized residual nucleic acids and β-actin PCR with and without the RT step to detect DNA).
See figures
Principle
Procedure
Applications
Supporting data and figures
Linearity over a broad dilution range.
Specifications
| Features | Specifications |
|---|---|
| Applications | Next-generation sequencing, end-point RT-PCR, quantitative, real-time RT-PCR, array analysis |
| Format | Spin column |
| Yield | Varies (depends on sample type, size and RNA contents) |
| Sample amount | <5 mg tissue or 5 x 105 cells |
| Time per run | 30–40 minutes |
| Main sample type | Tissue, cells, low biomass samples |
| Processing | Manual |
| Technology | Silica technology, ultra-clean production |
| Elution volume | 10–20 µl |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA (> 200 nt, to enrich mRNA) |