QuantiFast SYBR® Green PCR Kit
For fast, real-time PCR and two-step qRT-PCR using SYBR Green
For fast, real-time PCR and two-step qRT-PCR using SYBR Green

Cat. No. / ID. / ID. 204056
The QuantiFast SYBR Green PCR Kit delivers fast and specific quantification of gDNA or cDNA targets by real-time PCR or two-step RT-PCR using SYBR Green I detection. Q-bond technology and an optimized, ready-to-use master mix enable shorter real-time PCR run times, not only on fast cyclers with short ramping times, but also on standard cyclers. The combination of a hot start and a unique PCR buffer system in the ready-to-use master mix ensures highly sensitive qPCR on any real-time cycler without the need for optimization. For convenience, the master mix can be stored at 2–8°C.
IMPORTANT NOTE: As announced earlier, the production of the QuantiFast kits has been discontinued since mid-2021. Hence, these products will be available only until stocks last. Visit the product page of the successor kit to view improved features or to request a trial kit.
For more information and FAQs on this transition, visit: www.qiagen.com/PCRresource.
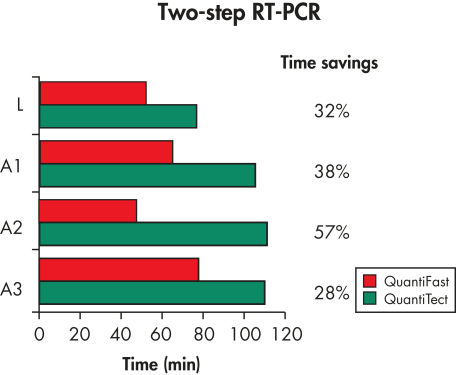
The QuantiFast SYBR Green PCR Kit delivers highly specific and sensitive results, outperforming other real-time PCR kits used in fast cycling mode (see figure " Sensitive two-step RT-PCR"). PCR run times are reduced by up to 60% (see figure " Significantly reduced PCR times"), allowing you to get results faster without compromising PCR performance (see figure " Faster results without compromising sensitivity"). You can also greatly increase your sample throughput or efficiently share a cycler with other users.
The QuantiFast SYBR Green PCR Kit enables accurate quantification of targets over several log dilutions of template. Even small differences in the amount of low-copy targets can be clearly distinguished (see figure " Resolution of small differences in copy number").
The QuantiFast SYBR Green PCR Kit delivers highly sensitive and specific results over a wide dynamic range on both standard and fast cyclers. The fluorescent dye SYBR Green I in the master mix enables the analysis of many different targets without having to synthesize target-specific labeled probes. A specially developed fast PCR buffer contains the novel PCR additive Q-Bond, which significantly reduces denaturation, annealing, and extension times (see figure " Fast primer annealing"). A balanced combination of K+ and NH4+ ions in the PCR buffer promotes specific primer annealing and enables high PCR specificity and sensitivity (see figure " Specific primer annealing"). In addition, HotStarTaq Plus DNA Polymerase requires only 5 minutes at 95°C for activation and provides a stringent hot start, preventing the formation of nonspecific products.
| Component | Features and benefits | Benefits |
| HotStarTaq Plus DNA Polymerase | 5 min activation at 95ºC | Set up of qPCR reactions at room temperature |
| QuantiFast SYBR Green PCR Buffer | Balanced combination of NH4+ and K+ ions | Specific primer annealing ensures reliable PCR results |
| Unique Q-Bond additive | Faster PCR run times, enabling faster results and more reactions per day | |
| SYBR Green I dye | Yields a strong fluorescent signal upon binding to double-stranded DNA | Highly sensitive amplification |
| ROX dye | Normalizes fluorescent signals on Applied Biosystems and, optionally, Agilent instruments | Precise quantification on cyclers that require ROX dye. Does not interfere with PCR on any real-time cycler |
The QuantiFast SYBR Green PCR Kit is a ready-to-use master mix that eliminates the need for optimization of reaction and cycling conditions. Simply add primers and DNA template to the ready-to-use PCR master mix, and start the reaction. Follow the protocol in the handbook to get fast and reliable results on any real-time cycler.
For optimal results in real-time two-step RT-PCR, we recommend synthesizing cDNA using the QuantiTect Reverse Transcription Kit. The kit provides fast cDNA synthesis in just 20 minutes with integrated removal of genomic DNA contamination.
We also recommend QuantiTect Primer Assays for gene expression analysis using SYBR Green. QuantiTect Primer Assays are bioinformatically validated primer sets for any gene from human, mouse, rat, and many other species. Assays can be easily ordered online at the GeneGlobe Web portal.
The QuantiFast SYBR Green PCR Kit is for use in gene expression analysis of cDNA targets and quantitative gDNA analysis. QuantiFast SYBR Green PCR Kits are compatible with all available real-time cyclers, including instruments from Applied Biosystems, Bio-Rad, Cepheid, Eppendorf, Roche, and Agilent. For the Rotor-Gene Q and other Rotor-Gene cyclers, we recommend using the Rotor-Gene SYBR Green PCR Kit, which has been specially developed for fast cycling on these instruments.
| Features | Specifications |
|---|---|
| Applications | SYBR Green-based, real-time PCR, two-step RT-PCR |
| Real-time or endpoint | Real-time |
| Sample/target type | cDNA, DNA |
| With or without ROX | With ROX |
| SYBR Green I or sequence-specific probes | SYBR Green I |
| Thermal cycler | Applied Biosystems, Bio-Rad, Cepheid, QIAGEN, Eppendorf, Roche, and Agilent |
| With/without hotstart | With hotstart |
| Single or multiplex | Single |
| Reaction type | Real-time and two-step RT-PCR |