✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 28181
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- 即使用可能なDNAの回収率が最高95%
- 迅速で簡便な操作
- 簡単な3ステップで最大10 kbまでのDNAをクリーンアップ
Product Details
QIAquick 96 PCR Kitsは96ウェルプレート、バッファー、コレクションチューブで構成され、シリカメンブレンによりPCR 産物(100 bp以上)のハイスループット精製を実現します。10 kbまでのDNAを簡便で迅速な結合-洗浄-溶出ステップおよび60~80 μlの溶出液により精製できます(得られるDNA溶出液は40~60 μl)。本クリーンアップ操作はQIAquick 96 PCR BioRobot Kitを用いてBioRobotワークステーションで完全自動化が可能です。
Performance
See figures
Principle
QIAquick 96 Kitsでは、高塩濃度バッファーによりDNAを結合し、低塩濃度バッファーあるいは水によりDNAを溶出するシリカメンブレンを利用しています。この精製法でDNAサンプルからプライマー、ヌクレオチド、酵素、ミネラルオイル、塩、アガロース、臭化エチジウム、およびその他の夾雑物が除去できます。シリカメンブレンテクノロジーにより樹脂漏れ、および懸濁液関連の問題や不便さは解消されます。それぞれの用途に応じて至適化された結合バッファーにより、種々の大きさのDNAフラグメントを選択的に吸着します。
QIAquick 96操作法は、96個までのPCRサンプルをQIAvac 96を用いた吸引法で効率的に精製します 。
また、QIAquick 96 PCR BioRobot Kit は、BioRobot 9600(販売終了)、BioRobot 3000(販売終了)およびBioRobot 8000での使用に最適化された特別なキットフォーマットです。本キットは、96個までのPCRサンプルの自動化ハイスループット・クリーンアップのために必要なすべてのバッファーとプラスチック容器が含まれたQIAquick 96モジュールとなっています。
Procedure
QIAquickシステムでは結合-洗浄-溶出という簡単な操作だけでDNAのクリーンアップが可能です(フローチャート" QIAquick 96操作手順"参照)。結合バッファーをPCRサンプルあるいは他の酵素反応液に直接添加し、混合液を96ウェルプレートにアプライします。DNAは添付のバッファー中の高塩濃度の条件下でシリカゲルメンブレンに吸着します。不純物は洗い流され、純粋なDNAを添付の低塩濃度溶出バッファー、あるいは水で溶出します。 得られたDNAは、様々なアプリケーションに即使用可能です。
操作法
QIAquickマルチウェルモジュール では、QIAvac装置を用いた吸引操作で精製を行なえます。QIAquick 96 PCR Purification Kits では、QIAvac 96吸引マニホールド(Cat. no. 9014579)を使用する必要があります。本クリーンアップ操作はQIAquick 96 PCR BioRobot Kitを用いてBioRobotワークステーションで完全自動化が可能です。
See figures
Applications
MinEluteまたはQIAquickシステムで精製したDNAフラグメントは、シークエンシング、マイクロアレイ解析、ライゲーション、トランスフォーメーション、制限酵素解析、標識反応、マイクロインジェクション、PCR、in vitro転写反応などのアプリケーションに即使用可能です。
Supporting data and figures
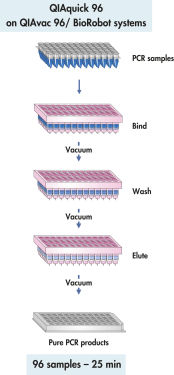
QIAquick 96 procedure.
The QIAquick 96 Kit uses a bind-wash-elute procedure with QIAvac 96 or the BioRobot Universal System.
Specifications
| Features | Specifications |
|---|---|
| Binding capacity | 10 µg |
| Processing | Manual/automated |
| Removal <10mers 17–40mers dye terminator proteins | Removal <40mers |
| Sample type: applications | DNA, oligonucleotides: PCR reactions |
| Format | 96-well plate |
| Fragment size | 100 bp – 10 kb |
| Technology | Silica technology |
| Recovery: oligonucleotides dsDNA | Recovery: oligonucleotides, dsDNA |
| Elution volume | 60–80 µl |