✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAseq Multimodal HC Panel HT (12)
Cat. No. / ID: 334942
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- QIAseq enrichment technology is the basis for Multimodal's uniform and complete coverage of targets, including complex genes like CEPBA, and confident detection of known and novel fusions
- Pre-optimized protocol to go from total nucleic acid extraction to preparation of UMI-containing, unique dual-indexed, NGS-ready libraries using a single-day workflow to enhance sensitivity, improve error correction and reduce index hopping
- Pre-configured automation-compatible workflow solution with secondary analysis and variant interpretation for rapid onboarding, faster turnaround time and improved scalability
Product Details
QIAseq Multimodal HT Panels have been developed for consolidated targeted DNA and RNA enrichment and analyses. Unlike other available approaches, QIAseq Multimodal HT Panels do not require 2 separate workflows for DNA and RNA analysis – saving time and conserving samples that are of limited availability.
Principle
Recent advances in NGS chemistries, platforms, and bioinformatics pipelines are enabling users to efficiently interrogate biological samples for changes in DNA and RNA. Other available approaches, however, require the use of 2 separate workflows to prepare libraries from separate DNA and RNA isolates. Limitations of such approaches include:
- Large amounts of sample material required for generating sufficient amounts of input DNA and RNA for multiple workflows
- Added complexity of deriving integrated insights from results of different technical approaches, each with its own innate bias
- Inefficient use of resources
- Long turn-around times
To overcome the limitations associated with current approaches, QIAseq Multimodal HT Panels start with total nucleic acids (or DNA + RNA) and prepares UDI-containing, Illumina-compatible targeted DNA and RNA libraries using a single-day, consolidated workflow. In addition, QIAseq Multimodal HT Panels have been designed for use with low-yield and poor-quality biological samples.
Procedure
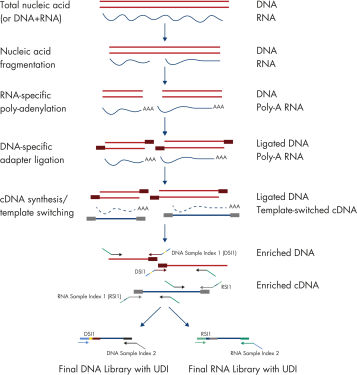
Workflow of the QIAseq Multimodal HT Panels
The QIAseq Multimodal HT Panel workflow can be used to prepare sequencing-ready libraries in a single day. The library insert size is approximately 150 bp, making the QIAseq Multimodal HT Panels highly compatible with low-quality samples, such as FFPE samples.
Robust detection of DNA and RNA biomarkers
QIAseq Multimodal HT Panels can be used to reliably detect DNA and RNA biomarkers using a consolidated workflow from total nucleic acids. The ability of QIAseq Multimodal HT Panels to simultaneously detect DNA and RNA biomarkers has been benchmarked against two separate workflows (namely, QIAseq Targeted DNA Pro and QIAseq Targeted Fusion XP).
Applications
QIAseq Multimodal HT Panels can be used to interrogate different types of biomarkers using a consolidated workflow from total nucleic acids, which can be isolated using dedicated sample isolation protocols that have been developed specifically for the QIAseq Multimodal HT Panels.
From DNA:
- Single nucleotide variants (SNVs)
- Insertions and Deletions (InDels)
- Copy number variants (CNVs)
- TMB score
- MSI status
From RNA:
- Fusions
- Exon skipping events
- Gene expression levels
Supporting data and figures
Workflow
The QIAseq Multimodal HT Panel workflow