QIAseq RNA Fusion XP Custom Panel (96)
Cat no. / ID. 334625
Features
- Incorporates QIAseq single primer extension (SPE) chemistry for design flexibility
- Unique molecular indices (UMIs) remove PCR duplicates and maximize sensitivity and specificity
- Fully customizable
- Streamlined workflow and single-tube enrichment pool for easy setup
- Excellent solution for difficult sample types such as FFPE
Product Details
QIAseq RNA Fusion XP enables the combined analysis of RNA fusion with SNV and gene expression from a single sample, allowing you to gain the complex RNA-seq insights your research demands. QIAGEN has leveraged our expertly curated design content and unparalleled custom assay capabilities to provide unprecedented NGS flexibility. Many other fusion detection systems suffer from a high false-positive (FP) rate, resulting from the high sensitivity these analyses demand. However, QIAseq Fusion XP Targeted Panels stand out among the rest and provide both an extremely low FP rate and high fusion detection sensitivity.
Incorporating our SPE chemistry, which requires just one gene-specific primer per target, QIAseq Fusion XP Targeted panels offer impressive flexibility for targeting fusions and for dense overlap of SNV regions of interest. Plus, all kits are optimized for use with our unique dual index (UDIs) adapters for use with Illumina® sequencing systems. Panels are fully customizable and support both gene-level and transcript-based designs, and our collection of catalogued panels includes focused targets for sarcoma, leukemia, lymphoma and other oncology applications.
Performance
The QIAseq RNA Fusion XP Panels deliver outstanding efficiency and sensitivity. An analysis using only 5 ng of control RNA as the input resulted in the successful detection of eighteen clinically relevant RNA gene fusions (see figure “ Outstanding efficiency and sensitivity”).
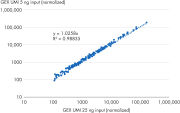
Relative gene expression quantitation by NGS is highly influenced by the read count, and even with the use of unique molecular indices (UMIs), it will still be affected by the amount of input RNA, the sequencing read depth and other factors. But the QIAseq Fusion XP Targeted Panels enable robust gene expression analysis. The built-in internal reference control make it possible to normalize the expression levels for various amounts of sample input (see figure “ High correlation gene expression over a 5-fold difference in RNA input”).
The QIAseq RNA Fusion XP Panels also provide outstanding sensitivity for detection of SNP/InDels. When used to analyze GM12878 RNA samples, the panels achieved 93.4% sensitivity for SNPs (VMF >=5%) within the covered and expressed target region. Furthermore, since cancer-specific markers often reside within exosomes, the panels can also detect SNPs in exoRNA (see figure “ High sensitivity SNP/InDel detection”).
See figures
Procedure
Isolated total RNA (10 ng or even less), is reverse transcribed with high efficiency into first-strand cDNA, followed by a robust second-strand cDNA synthesis. In the subsequent library construction step, adaptor complexes containing unique molecular indices (UMIs) and sample indices are incorporated into the dsDNA generated in the previous step. Next, QIAseq’s SPE chemistry is used to enrich the targets of interest, which can be any combination of gene fusions, gene expression and SNP/InDel targets. Finally, the libraries are amplified via fast universal PCR, in which a second index is added (dual indexing).
Applications
QIAseq RNA Fusion XP Panels use single primer extension (SPE) and unique molecular index (UMI) technologies in NGS to help identify and characterize fusion gene events, gene expression, and SNP/InDel at the RNA level.
Supporting data and figures
High sensitivity SNP/InDel detection
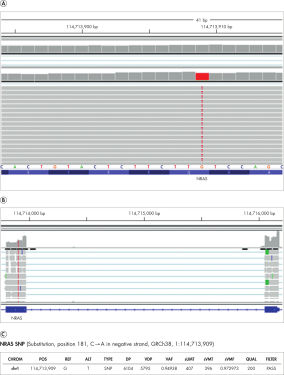
Using GM12878 RNA samples, QIAseq Fusion XP Targeted Panels achieved 93.4% sensitivity for SNPs (VMF >=5%) within the covered and expressed target region. Further, since cancer-specific markers often reside within exosomes it can also detect SNPs in exoRNA as shown.
A IGV browser illustrating the detection of an NRAS SNP from an exoRNA control that harbors the NRAS SNP (Substitution, position 181, C➞A in negative strand, GRCh38, 1:114713909). B A neighboring exon-exon junction region signal confirms the change is present at the RNA level. C Summary of the SNP call.