✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy MinElute Cleanup Kit (50)
Cat. No. / ID: 74204
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Product Details
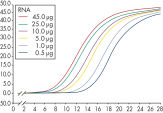
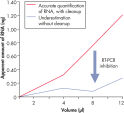
The RNeasy MinElute Cleanup Kit enables cleanup and concentration of RNA from enzymatic reactions or other samples using specialized RNeasy MinElute spin columns based on silica-membrane technology. The kit can also be used to desalt RNA samples. Up to 45 µg RNA can be purified in a volume as low as 10 µl. Purification can be fully automated on the QIAcube Connect.
Performance
See figures
Principle
The RNeasy MinElute Cleanup Kit is designed to purify and concentrate RNA from enzymatic reactions (e.g., labeling, in vitro transcription), RNA isolated by alcohol-precipitation and organic-extraction methods, as well as to desalt RNA samples, and concentrate RNA prepared by silica-membrane procedures (e.g., PAXgene Blood RNA preps). RNeasy technology simplifies total RNA isolation by combining guanidine-isothiocyanate lysis with silica-membrane purification.
Procedure
Guanidine-isothiocyanate–containing lysis buffer and ethanol are added to the sample to create conditions that promote selective binding of RNA to the RNeasy MinElute membrane. The sample is then applied to the RNeasy MinElute spin column. RNA binds to the silica-membrane, contaminants are efficiently washed away, and high-quality RNA is eluted in water.
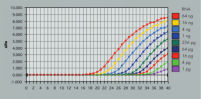
Enzymatic reactions or crude RNA preps are simultaneously cleaned up and concentrated in less than 15 minutes. In comparison, time-consuming concentration and cleanup by alcohol precipitation can result in loss of RNA, especially from small samples. The RNeasy MinElute procedure is faster than vacuum centrifugation, which only concentrates the sample without removing salts and other impurities. The unique design of RNeasy MinElute spin columns enables concentration of purified RNA to as little as 10 µl for downstream applications such as microarray analysis and real-time RT-PCR. Purification can be fully automated on the QIAcube Connect.
Applications
RNA purified with RNeasy MinElute technology is high-quality and ideal for use in all applications including:
- Northern, dot, and slot blotting
- End-point RT-PCR
- Quantitative, real-time RT-PCR
- Array analysis
- Poly A+ RNA selection
Supporting data and figures
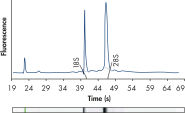
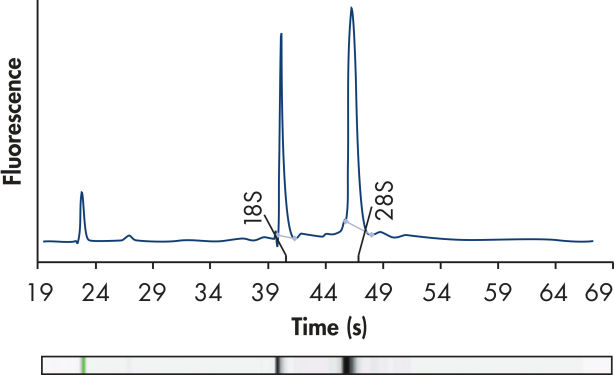
High-quality RNA.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, qPCR, real-time PCR, microarray |
| Format | MinElute Spin column |
| Sample amount | 200 µl |
| Elution volume | 10–14 µl |
| Processing | Manual |
| Main sample type | (Crude) RNA preps |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA |
| Time per run or per prep | <15 minutes |
| Technology | Silica technology |
| Yield | 45 µg |