✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
DNeasy Plant Pro Kit (50)
Cat. No. / ID: 69204
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Superior PCR performance with patented Inhibitor Removal Technology (IRT)
- Rapid extraction of ready-to-use DNA
- No organic extraction, no ethanol precipitation
- Highly efficient lysis and release of DNA from tough plant materials and associated plant pathogens
Product Details
The DNeasy Plant Pro Kit has several innovative features to overcome the challenges of DNA extraction from plant tissue and enable recovery of high-quality DNA from the toughest sample types, including strawberry leaf, grapevine leaf, pine needles and various seed types. Increased yields of plant DNA and plant pathogen DNA combined with superior removal of inhibitors ensure high-performance results in sensitive downstream applications.
Want to try the DNeasy Plant Pro Kit for the first time? Request a trial kit quote.
DNeasy Plant Kits provide fast and easy silica-based DNA isolation from plant samples in spin column format. Typical yields are 3–260 μg of high-quality DNA, depending on the samples used (e.g., wheat, maize, Arabidopsis, tomato, tobacco) and binding capacity of the DNeasy silica membrane. DNeasy Plant Kits also provide silica-based DNA purification in a convenient 96-well plate format with typical yields of 1–15 µg DNA, depending on the plant species. DNA isolation from plant tissue is also automatable on the QIAcube Connect.
Performance
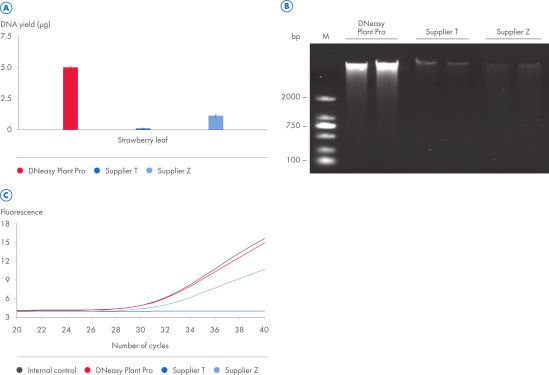
The DNeasy Plant Pro Kit – the newest member of the trusted DNeasy Plant family – enables purification of significantly higher yields of DNA from the toughest sample types, including strawberry leaf, grapevine leaf, pine needles and various seed types (see figure “ Significantly higher yields of pure DNA”). Patented Inhibitor Removal Technology (IRT) provides superior removal of inhibitors without using harsh chemicals, yielding pure DNA with less PCR inhibition (see figure “ Efficient removal of PCR inhibitors”).
When CT values of PCR reactions with plant DNA eluates containing possible inhibitors were compared to CT values of the PCR reaction with water added as control which does not inhibit amplification of the IC DNA, the eluate from the DNeasy Plant Pro Kit showed no inhibition.
With the DNeasy Plant Pro Kit, yields of DNA purified from strawberry leaf – a particularly tough plant sample – were significantly higher than those obtained using a kit from Supplier T and Supplier Z. Furthermore, the DNeasy Plant Pro Kit provided superior removal of inhibitors and the DNA eluate showed no inhibition (see figure “ Significantly higher yields and superior inhibitor removal”). The DNeasy Plant Pro Kit can also purify bacterial, fungal and viral DNA from plant and root samples.
The kit can be successfully combined into a workflow with proven products optimized for next-generation sequencing (NGS) (see figure “ Optimized NGS workflow”). Rapid and reliable identification of plant pathogens is crucial to avoid disease spread and facilitate effective disease management. Plant and pathogen DNA co-purified using the DNeasy Plant Pro Kit enables successful identification of a range of pathogens (see figure “ Successful identification of plant pathogens by NGS”).
DNeasy Plant Kits allow rapid and efficient isolation of high-quality DNA from a wide variety of plant species and tissue types, including the most demanding sources (see table "Selection of plant species processed with DNeasy Kits"). Samples may be fresh, frozen or dried. The optimized DNeasy Plant procedure incorporates the QIAshredder spin column, a unique filtration and homogenization column that efficiently removes cell debris and improves sample handling following lysis.
The typical yield is 3–260 µg, with a sample size of up to 1 g wet weight, and an elution volume of 50 µl to 2 ml. DNA yields vary between different species and tissues depending on genome size, ploidy, cell number and tissue sample age.
| Abies alba (silver fir) | Nicotiana tabacum (tobacco) |
| Aesculus hippocastanum (horse chestnut) | Oryza sativa (rice)4 |
| Arabidopsis thaliana (thale cress) | Pelargonium sp. (geranium)4 |
| Avena sp. (oat) | Petunia sp.4 |
| Brassica napus (oilseed rape) | Pinus sylvestris (Scotch pine), P. brutia5 |
| Brassica oleracea (kohlrabi) | Populus tremula (aspen) |
| Chicorium endivia (chicory) | Pseudotsuga menziesii (Douglas fir) |
| Citrullini lanatus (water melon) | Quercus robur, Q. petrea (oak)6,7 |
| Egeria sp. | Rhododendron sp.2,4 |
| Fagus sylvatica (beech)1 | Rubus idaeus (raspberry) |
| Helianthus spp. (sunflower) | Solanum tuberosum (potato) |
| Hordeum vulgare (barley)2 | Sphagnum palustre (moss) |
| Humulus sp. (hops) | Spinacia oleracea (spinach) |
| Hydrilla sp. | Taxus baccata (yew) |
| Kalanchoe spp. | Triticum aestivum (wheat)4 |
| Lupinus sp. | Ulmus glabra (elm)6 |
| Lycopersicon esculentum (tomato)3 | Vitis spp. (grape)6 |
| Myriophyllum sp. | Zea mays (maize) |
The DNeasy Plant procedure yields pure nucleic acid, free of polysaccharides and other secondary metabolites often copurified using conventional methods. Such impurities can interfere with spectrophotometric readings and inhibit enzymatic reactions. Absorbance scans of DNeasy purified DNA show a symmetrical peak at 260 nm (see figures " DNA purity from oak leaves and pine needles"), confirming that the DNA is free of impurities, including enzyme inhibitors. DNeasy purified DNA is sized up to 40 kb (see figure " Pure DNA (20–25 kb) for restriction analysis"). Purified DNA can be used in a wide range of applications (see figures " PCR analysis" and " RAPD analysis").
High-quality DNA can be isolated from 96 or 192 samples of plant leaf tissue in less than 2 hours. The simple 96-well procedure provides highly reproducible yields of total cellular DNA (see figure " Uniformity of DNA yields from 96 wheat samples"). The typical yield is 1–15 µg per 50 mg plant leaf tissue, with an elution volume of 100–200 µl. DNA yields vary between species depending on genome size, ploidy, cell number and tissue sample age.
See figures
 Efficient removal of PCR inhibitors.
Efficient removal of PCR inhibitors.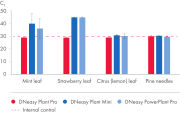 Optimized NGS workflow.
Optimized NGS workflow. Successful identification of plant pathogens by NGS.
Successful identification of plant pathogens by NGS.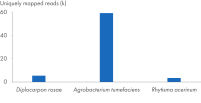 DNA purity from oak leaves and pine needles.
DNA purity from oak leaves and pine needles. Pure DNA (20–25 kb) for restriction analysis.
Pure DNA (20–25 kb) for restriction analysis.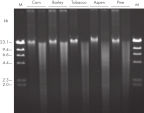 PCR analysis.
PCR analysis. RAPD analysis.
RAPD analysis.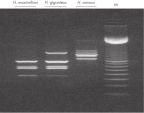 Uniformity of DNA yields from 96 wheat samples.
Uniformity of DNA yields from 96 wheat samples.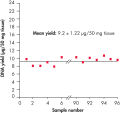
Procedure
The DNeasy Plant Pro Kit uses bead-beating technology, which replaces cumbersome DNA isolation procedures such as CTAB, phenol or chloroform extraction to recover high-quality DNA from the toughest sample types, including strawberry leaf, grapevine leaf, pine needles and various seed types. The DNeasy Plant Pro Kit uses the second generation of QIAGEN’s patented Inhibitor Removal Technology (IRT) to remove PCR inhibitors, including polysaccharides, polyphenolics and other secondary metabolites from plant extracts during the isolation process. Improved IRT, combined with efficient bead beating and lysis chemistry, results in high yields of inhibitor-free DNA that is ready for immediate use in downstream applications, including PCR, qPCR, and RAPD analysis, RFLP analysis, Southern blotting and next-generation sequencing.
Samples are added to the Tissue Disruption Tube which contains a specially shaped bead and a buffer for rapid homogenization (see figure " Rapid homogenization in Tissue Disruption Tubes"). Cell lysis and DNA release occur by mechanical and chemical methods. Released genomic DNA is cleared of PCR inhibitors using QIAGEN’s second-generation Inhibitor Removal Technology (IRT) and then captured on a silica membrane in a spin column format. DNA is then washed and eluted from the membrane and is ready for PCR, qPCR, NGS and other downstream applications.
An optional Phenolic Separation Solution step helps provide pure DNA even from samples with high polyphenolic compounds, such as pine or grape leaves by breaking the bonds between DNA and phenolics. This step prevents DNA loss that would otherwise occur in these samples.
The DNeasy Plant Pro Kit is used with a vortexer or high-velocity bead beating, based on sample needs. Vortex methods work with soft leaf tissue. High-velocity bead beaters, like the PowerLyzer 24 Homogenizer or the TissueLyser II, break down tougher plant material such as roots, seeds, stems or challenging leaf tissues. Furthermore, purification of DNA using the DNeasy Plant Pro Kit can be fully automated on the QIAcube Connect.
Samples are first mechanically disrupted and then chemically lysed (see flowchart " DNeasy Plant and DNeasy 96 Plant procedures"). RNA is removed by RNase digestion during lysis. Cell debris, precipitated proteins and polysaccharides are removed and the sample is homogenized by centrifugation through a QIAshredder spin column. Buffering conditions are adjusted and the lysate is loaded onto a DNeasy Plant spin column or 96-well plate. During a brief spin, DNA selectively binds to the silica membrane while contaminants pass through. The remaining contaminants and enzyme inhibitors are removed in one or two efficient wash steps. Pure DNA is then eluted in water or low-salt buffer, ready for use.
Purification of DNA using the DNeasy Plant Mini Kit can be automated on the QIAcube Connect.
See figures
Applications
DNeasy Plant Kits provide purification of ready-to-use DNA from plant samples, including plant cells, plant tissues and fungi.
The DNeasy Plant Pro Kit is designed for:
- DNA extraction from plants and plant-associated microorganisms
- PCR and NGS analysis
- Marker-assisted breeding
- Plant pathogen research
- Studies on genetically modified plants
- Detection of resistance traits
| Features | DNeasy Plant Mini Kit | DNeasy Plant Maxi Kit | DNAeasy 96 Plant Kit | DNeasy Plant Pro Kit |
|---|---|---|---|---|
| Applications | PCR, qPCR, blotting, next-generation sequencing | PCR, qPCR, blotting, next-generation sequencing | PCR, qPCR, blotting, next-generation sequencing | PCR, qPCR, blotting, next-generation sequencing |
| Elution volume | 50–200 µl | 500 µl – 2 ml | 100–200 µl | 50–100 µl |
| Format | Spin column | Spin column | 96-well plate | Spin column |
| Main sample type | Plant samples | Plant samples | Plant samples | Plant samples and seeds |
| Bead size | N.A. | N.A. | N.A. | 5/32” (3.9 mm) ballcone |
| Binding capacity | Up to 50 µg | Up to 500 µg | Up to 50 µg (per well) | Up to 50 µg |
| Processing | Manual | Manual | Manual | Bead beating |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | DNA | DNA | DNA | DNA |
| Sample amount | Up to 100 mg | Up to 1 g | Up to 50 mg | Up to 100 mg |
| Technology | Silica technology | Silica technology | Silica technology | Silica technology |
| Throughput | Varies | Varies | 96 or 192 samples | Varies |
| Time per run or per prep | <1 hour | <2 hours | <2 hours (192 samples) | 45 minutes (24 samples) |
| Typical yield (from 50 mg starting material) | Up to 30 µg | Up to 260 µg | Up to 15 µg | Up to 30 µg |
N.A. = Not applicable
Supporting data and figures
Significantly higher yields and superior inhibitor removal.