Features
- Compliance with EU IVD Directive 98/79/EC
- Comprehensive results in real time
- Accurate quantification of mutations in the KRAS gene
- Easy interpretation of complex sequence information
Product Details
Performance
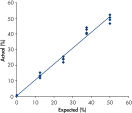
Linearity
Linearity was measured according to CLSI Guideline EP6-A “Evaluation of the linearity of quantitative measurement procedures: a statistical approach; approved guideline”.
Plasmids carrying wild type and mutant sequences were mixed in proportions to give the following levels of mutation: 0, 12.5, 25, 37.5, and 50%. Four replicates of the mixes were placed in a random pattern on a plate and analyzed. The results for the mutation GGT->TGT in codon 12 are shown (figure " Linearity of GGT->TGT mutation").
Precision
Determination of the linearity of the mutation GGT->TGT in codon 12 was repeated by 3 operators on 3 separate days using different combinations of PyroMark Q24 instruments and reagents.
The values for the intermediate precision (SD) were 0.6–2.0 % units in the measured range of 0–50% mutation level.
| % mutated plasmid | Run 1 (Mean, SD) | Run 2 (Mean, SD) | Run 3 (Mean, SD) | Summary (Mean, SD) |
|---|---|---|---|---|
| 0.0 | 0.6, 0.2 | 1.7, 0.7 | 0.7, 0.2 | 1.0, 0.6 |
| 12.5 | 13.3, 1.5 | 16.2, 1.9 | 14.6, 3.0 | 14.7, 1.4 |
| 25.0 | 23.8, 1.4 | 26.8, 2.4 | 26.9, 2.9 | 25.8, 1.8 |
| 37.5 | 42.0, 1.9 | 41.7, 0.5 | 38.5, 2.6 | 40.7, 2.0 |
| 50.0 | 49.7 2.4 | 50.5, 1.8 | 49.1, 4.8 | 49.8, 0.7 |
See figures
Principle
The therascreen KRAS Pyro Kit is used for quantitative measurements of mutations in codons 12, 13, and 61 of the human KRAS gene in real time using Pyrosequencing technology on the PyroMark Q24 System. The KRAS gene encodes a protein that plays a critical role in the EGFR signaling cascade. Mutations in the KRAS gene can affect how the protein stimulates these downstream pathways. KRAS is mutated in approximately 30% of all cancer types. Cancers that exhibit a high frequency of KRAS mutation include colorectal cancer (35%) and lung (18%) cancer.
The following mutations are detected:
- Codon 12/13: G12A, G12C, G12D, G12R, G12S, G12V, G13D
- Codon 61: Q61E, Q61H, Q61L, Q61R
Procedure
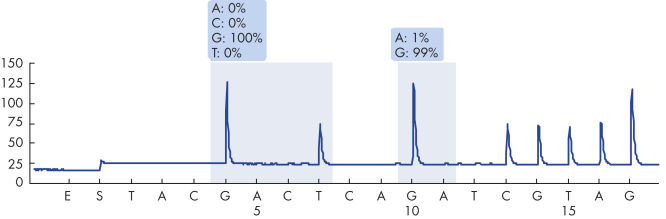
After PCR using primers targeting codons 12/13 and codon 61, the amplicons are immobilized on Streptavidin Sepharose High Performance beads. Single-stranded DNA is prepared, and the corresponding sequencing primers anneal to the DNA. The samples are then analyzed on the PyroMark Q24 System using a run setup file and a run file. The KRAS Plug-in Report should be used to analyze the run. This report ensures that the correct limit of detection (LOD) values are used and different sequences to analyze are automatically used to detect all mutations. However, the run can also be analyzed using the analysis tool integral to the PyroMark Q24 System (see figure " Pyrogram trace of a GGT to GAT mutation in base 2 of codon 12"). The "Sequence to Analyze" can be then adjusted for detection of rare mutations after the run (see figures " Pyrogram trace of a GGT to TGT mutation in base 1 of codon 12" and " Pyrogram trace of the reanalysis of the sample in the previous figure").
See figures
Applications
The following mutations are detected:
- Codon 12/13: G12A, G12C, G12D, G12R, G12S, G12V, G13D
- Codon 61: Q61E, Q61H, Q61L, Q61R
Supporting data and figures
Pyrogram trace of a normal genotype in codons 12 and 13.