✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAamp DNA Blood Mini Kit (50)
Cat. No. / ID: 51104
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Rapid purification of high-quality, ready-to-use DNA
- No organic extraction or alcohol precipitation
- Consistent, high yields
- Complete removal of contaminants and inhibitors for reliable results
- Kit formats for low- to high-throughput – options for automation of all kits
Product Details
QIAamp DNA Blood Kits provide silica-membrane-based DNA purification from whole blood, plasma, serum and other body fluids. The kits are designed for a range of sample sizes from 200 μl up to 10 ml fresh or frozen human whole blood. QIAamp spin columns can be easily processed in a centrifuge or on vacuum manifolds. A convenient 96-well format using centrifugation enables purification of DNA for labs that need high-throughput DNA purification from blood, buffy coat, plasma, serum, bone marrow, lymphocytes and body fluids. A dedicated kit is also available for automated purification of 1–12 samples on the QIAcube Connect.
Performance
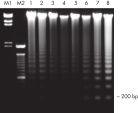
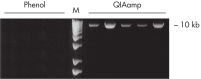
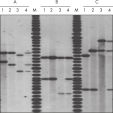
QIAamp DNA Blood Kits yield DNA sized from 200 bp to 50 kb, depending on the age and storage of samples (see figure "Apoptotic banding in stored blood"). The purified DNA is suitable for long-range PCR amplification (see figure " Long-range PCR") and restriction fragment length polymorphism (RFLP) analysis used, for example, for paternity testing (see figure " Paternity testing by RFLP analysis").
The QIAamp DNA Blood Midi Kit yields up to 94.5% recovery of DNA; the QIAamp DNA Blood Maxi Kit yields up to 95.8% recovery of DNA, depending on the starting cell densities (see table “DNA yields from human whole blood with different cell densities”).
| Leukocytes per ml | DNA yield (µg) | DNA recovery (%) | ||
| QIAamp Midi | QIAamp Maxi | QIAamp Midi | QIAamp Maxi | |
| 2.5 x 105 | 3.0 | 15.8 | 92.3 | 95.8 |
| 1.0 x 106 | 11.0 | 60.4 | 91.7 | 91.5 |
| 5.0 x 106 | 62.4 | 312.0 | 94.5 | 94.5 |
| 1.0 x 107 | 116.3 | 624.6 | 88.1 | 94.6 |
Genomic DNA was purified from 2 ml (QIAamp Midi) or 10 ml (QIAamp Maxi) human whole blood and eluted in 300 µl (Midi) or 1 ml (Maxi) elution buffer. The first eluate was loaded onto the column a second time and centrifuged again (i.e., re-eluted). Percentage DNA recovery was calculated by assuming that one leukocyte contains 6.6 pg DNA.
The QIAamp 96 DNA Blood Kit processes up to 200 µl sample size, with a preparation time of 192 samples in two to three hours, yielding highly pure DNA in less than one minute per preparation. Even higher throughput can be achieved by staggering the procedure. QIAamp 96 plates provide well-to-well uniformity in DNA recovery and purity (see figures " Sample reproducibility" and " Reproducibility of yield and purity"). The typical yield is 6 µg per 200 µl healthy whole blood, with an elution volume of 50–200 µl.
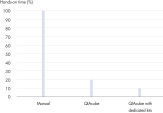
The dedicated QIAamp DNA Blood Mini QIAcube Kit enables automated DNA isolation from blood and DNA isolation from body fluids on the QIAcube Connect. The kit includes rotor adapters that are preloaded with QIAamp spin columns and elution tubes, delivering greater convenience and time savings (see figure " Significant time savings"). Furthermore, ease of use is increased and user errors minimized. Waste is reduced, because the content of the dedicated kit is tailored for purification on the QIAcube Connect and the superfluous tubes that are required for the manual procedure are not included.
If using the automatable QIAamp DNA Blood Mini Kit on the QIAcube Connect, the QIAamp DNA Blood Mini Accessory Set A and QIAamp DNA Blood Mini Accessory Set B provide the convenience of extra buffers and reagents for automated sample prep.
See figures
Principle
No phenol–chloroform extraction is required. DNA binds specifically to the QIAamp silica-gel membrane while contaminants pass through. PCR inhibitors, such as divalent cations and proteins, are completely removed in two efficient wash steps, leaving pure nucleic acid to be eluted in either water or a buffer provided with the kit. QIAamp DNA Blood technology yields genomic, mitochondrial or viral DNA from blood and related body fluids ready to use in PCR and blotting procedures. QIAamp sample preparation technology is fully licensed.
Procedure
QIAamp DNA Blood Kits simplify DNA purification from blood and DNA purification from body fluids with fast spin-column, vacuum, centrifugation or automated procedures (see figure “ QIAamp Spin Column procedure”). Fresh and frozen whole blood with common anticoagulants, such as citrate, EDTA and heparin, may be processed. If processing QIAamp Midi or Maxi spin columns on vacuum manifolds additional Buffer AW1 [cat. no. 19081] and Buffer AW2 [cat. no. 19072] are required for use with the vacuum manifolds.
Vacuum processing with the QIAamp DNA Blood Mini Kit
With the QIAamp DNA Blood Mini Kit, blood can be processed by vacuum instead of centrifugation, for greater speed and convenience in DNA purification. QIAamp mini spin columns are accommodated on the QIAvac 24 manifold using VacValves and VacConnectors. VacValves should be used if sample flow rates differ significantly, to ensure consistent vacuum. Disposable VacConnectors are used to avoid any cross-contamination. Use of VacConnectors also allows these QIAamp spin procedures to be performed on QIAvac 6S with QIAvac Luer Adapters.
Automated processing on the QIAcube Connect
The award-winning QIAcube Connect uses advanced technology to process QIAGEN spin columns, enabling seamless integration of automated, low-throughput sample prep into laboratory workflows. All steps in the purification procedure are fully automated – up to 12 samples can be processed per run. The QIAcube Connect used together with the dedicated QIAamp DNA Mini QIAcube Kit provides a winning combination for fast, easy and convenient DNA purification.
High-throughput processing in 96-well format
The QIAamp 96 spin procedure requires the QIAGEN 96-Well-Plate Centrifugation System’s deep rotor buckets to accommodate QIAamp 96 plates stacked on 96-well blocks. Fresh or frozen whole blood treated with common anticoagulants such as, EDTA, citrate and heparin, may be used. Dried whole blood may be processed with additional equipment and the use of a special protocol available from QIAGEN Technical Services or your local distributor.
See figures
Applications
QIAamp DNA Blood Kits provide proven QIAamp technology for the purification of DNA from a variety of materials. Sample sources include:
- Fresh and frozen whole blood or buffy coat
- Plasma or serum
- Bone marrow
- Lymphocytes
- Platelets
- Body fluids
- Cultured cells
- Swabs and buccal cells
| Features | QIAamp DNA Blood Mini Kit | QIAamp DNA Blood Midi Kit | QIAamp DNA Blood Maxi Kit | QIAamp 96 DNA Blood Kit |
| Applications | PCR, long-range PCR, Southern blotting | PCR, Southern blotting | PCR, blotting | Southern blotting, genome mapping |
| Elution volume | 50–200 µl | 100–400 µl | 500–2000 µl | 50–200 µl |
| Format | Spin column | Spin column | Spin column | 96-well plate |
| Main sample type | Whole blood, body fluids | Whole blood, body fluids | Whole blood, body fluids | Whole blood, body fluids |
| Processing | Manual (centrifugation or vacuum) | Manual (centrifugation or vacuum) | Manual (centrifugation or vacuum) | Manual (centrifugation or vacuum) |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | Genomic DNA, mitochondrial DNA, viral DNA | Genomic DNA, mitochondrial DNA, viral DNA | Genomic DNA, mitochondrial DNA, viral DNA | Genomic DNA, mitochondrial DNA, viral DNA |
| Sample amount | 1–200 µl | 0.3–2 ml | 3–10 ml | <200 µl |
| Technology | Silica technology | Silica technology | Silica technology | Silica technology |
| Time per run or per prep | 20–40 minutes | 55 minutes | 55 minutes | 2–3 hours (192 samples) |
| Yield | 4–12 µg | 20–60 µg | 300–600 µg | 6 µg |
Supporting data and figures
Exemplary size distribution using QIAamp DNA Blood Mini Kit (Blood)
Exemplary size distribution of DNA from blood using the QIAamp DNA Blood Mini Kit. Samples were analyzed with the Femto Pulse System and signals were normalized to maximum peak.