✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
REPLI-g Mini Kit (25)
Cat no. / ID. 150023
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Amplified DNA can be used in NGS, microarray and PCR applications
- Isothermal multiple displacement amplification (MDA) with proof-reading DNA polymerase
- Whole genome amplification (WGA) yields 7 to 40 µg amplified DNA from 10 ng of DNA
- Ultrafast format for whole genome amplification in 1–1.5 hours
- High-throughput format for 96 samples with just 20 minutes of handling time
Product Details
Limitations on the number and type of genomic analyses that can be performed due to insufficient quantities of genomic DNA can be overcome by global amplification of all DNA within a sample (whole genome amplification). REPLI‑g Kits provide DNA polymerase, buffers and reagents for highly uniform whole genome amplification with minimal sequence bias and can be used with various starting materials, including genomic DNA, fresh or dried blood, buccal swabs, fresh or frozen tissue and cells. QIAGEN REPLI-g Kits are available in several different sizes and configurations which allows researchers to build the best workflow for their application and lab space. Typical REPLI-g yields range from approx. 7 µg to 40 µg of amplified DNA, depending on which kit and workflow is used.
The REPLI-g Mini and Midi Kits offer a single tube workflow with higher yields of amplified DNA, while the REPLI-g UltraFast Kit enables highly uniform and accurate whole genome amplification in just 60–90 minutes. For high-throughput processing in 96-well format, the streamlined, room-temperature procedure of the REPLI-g Screening Kit enables straightforward liquid handling that requires just 20 minutes. The REPLI-g Screening kit is intended for rapid screening using a range of genotyping, PCR, sequencing or microarray assays.
Performance
High yields from a variety of samples, suitable for numerous applications
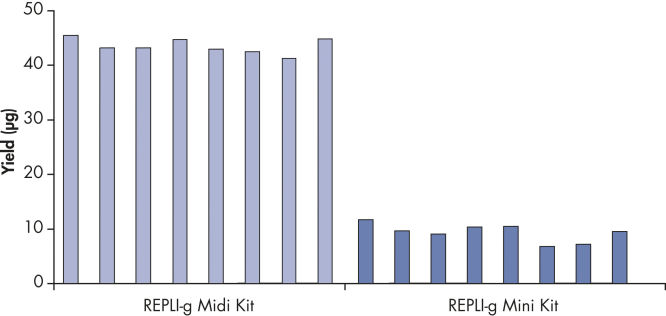
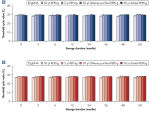
Various clinical and non-clinical research samples can be used with REPLI-g Mini and Midi Kits, including genomic DNA, fresh or dried blood, fresh or frozen tissue and cells. Typical DNA yields per 50 µl reaction consistently reach 10 µg (mini kit) or 40 µg (midi kit) (see figure " Consistent DNA yields using any sample type"), while a uniform yield of amplified DNA is usually achieved regardless of the quantity of template DNA (see figure " Uniform DNA yield from various amounts of template"). Obtaining uniform DNA yields from varying template concentrations is always important, but particularly essential for high-throughput applications, which require subsequent genetic analyses to be possible without additional measurement or adjustment of DNA concentration.
The average DNA yield from the REPLI-g UltraFast Mini Kit is 7–10 µg per 20 µl reaction in 90 minutes with standard quality samples. If time is very limited, sufficient product is available for downstream genetic analysis after just 45 minutes (see figure " Consistent and high yields of amplified DNA"). Negative control samples (without template DNA) are not amplified, providing a reliable indicator of the presence/absence of sample or contaminating DNA (see figure " Consistent and high yields of amplified DNA").
Typical DNA yields from a REPLI-g Screening Kit reaction are 8 µg per 40 µl reaction. The amplified DNA can be used directly in a variety of downstream applications, including genotyping (e.g., SNP, STR, deletions and insertions), end-point PCR, quantitative real-time PCR, microarray and sequencing. Unlike other whole genome amplification procedures that require the reaction setup to be performed on ice, the REPLI-g Screening Kit uses room temperature setup with just 20 minutes of handling time, making it especially suited for processing multiple samples in parallel. Obtaining uniform DNA yields from varying template concentrations is particularly important for high-throughput applications to allow subsequent genetic analysis without the need to measure or adjust DNA concentration. The average product length is typically greater than 10 kb, with a range between 2 kb and 100 kb (see figure “ Uniform yield of high-molecular-weight DNA”).
The average product length of REPLI-g amplified DNA is typically more than 10 kb, with a range between 2 kb and 100 kb, enabling downstream applications such as complex restriction enzyme analysis and long-range PCR to be carried out. REPLI-g amplified DNA is highly suited for genotyping applications, such as SNP genotyping with TaqMan® primer/probe sets (see figure " Reliable SNP genotyping"), sequencing and STR/microsatellite analysis (see figure " Accurate genotyping").
Researcher feedback
"In the past two years, we have been using the REPLI-g Ultra Fast Mini Kit extensively for multiple projects. The multiple displacement amplification performed with the REPLI-g Kit was able to faithfully amplify all DNA products at very low error rate and high coverage. The downstream applications we have used the kit for include most often NGS, but also qPCR and microarray. Moreover, we are working on implementing this technology in microfluidic or lab-on-a-chip systems, since the method requires no sophisticated thermal cycling program." Dr. Rong Fan, Assistant Professor of Biomedical Engineering, Yale University, US.
Successfully used in next-generation sequencing
Numerous publications have demonstrated the successful utilization of REPLI-g amplified DNA for next-generation sequencing (NGS) applications that range from exome and whole genome sequencing of tumor cells, to metagenomics research, to single cell analysis (for a range of recent publications that successfully used REPLI-g in NGS, please see our WGA resource page). Since the use of whole genome amplified DNA for NGS and array applications has been debated, we detected potential factors that could influence the success of using amplified DNA for these downstream applications. We determined that the quality of input material strongly influences the success of downstream NGS experiments. If working with low quality DNA (e.g., degraded DNA) or aged tissue material, the resulting amplified DNA may not give reliable results (data not shown). However, WGA, using REPLI-g technology, on intact cells or non-degraded purified DNA shows that NGS results are comparable to those obtained with purified gDNA. Sequence coverage and alignment comparison of the genomic loci sequence indicates minimized levels of junk DNA after WGA, whereas error rates are in a similar percentage range for both amplified and genomic DNA (see figure “ Comparable NGS (next-generation sequencing) results obtained using purified gDNA or REPLI-g amplified DNA”). The QIAseq FX Single Cell RNA and QIAseq FX Single Cell DNA Library Kits utilize the REPLI-g MDA reaction for high quality NGS libraries when starting with 10 pg – 1 ng of RNA or DNA.
High fidelity whole genome amplification
REPLI-g technology provides highly uniform DNA amplification across the entire genome. Phi29 polymerase can replicate up to 70 kb without dissociating from the genomic DNA template (see figure " Schematic representation of REPLI-g amplification"). In contrast to PCR-based whole genome amplification (WGA) technologies, Phi29 polymerase has 3'→5' exonuclease proofreading activity and maintains up to 1000-fold higher fidelity compared to Taq DNA polymerase during replication. Exonuclease-resistant primers provided in the kit ensure high yields of DNA product, and the WGA buffer system is optimized for very long read length and unbiased locus representation.
REPLI-g outperforms PCR-based WGA methods
Traditional methods of genomic DNA amplification include the time-consuming process of creating EBV-transformed cell lines followed by whole genome amplification using random or degenerate oligonucleotide-primed PCR. Also, PCR-based methods (e.g., DOP-PCR and PEP), as generally used by other suppliers, can produce nonspecific amplification artifacts and give incomplete coverage of loci. In several cases, DNA less than 1 kb long may be generated that cannot be used in many downstream applications. In general, the resulting DNA is generated with a much higher mutation rate due to the use of the low-fidelity enzyme Taq DNA polymerase, which can lead to error-prone amplification that results in, for example, single base-pair mutations, STR contractions and expansions. In contrast to these disadvantages, REPLI-g provides highly uniform amplification across the entire genome, with minimal locus bias and minimized mutation rates during amplification (see figures " Highly representative amplification using REPLI-g technology" and " Consistent and accurate whole genome amplification").
See figures
 Uniform DNA yield from various amounts of template.
Uniform DNA yield from various amounts of template.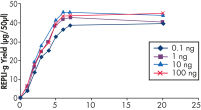 Consistent and high yields of amplified DNA.
Consistent and high yields of amplified DNA.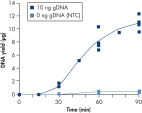 Uniform yield of high-molecular-weight DNA.
Uniform yield of high-molecular-weight DNA. Reliable SNP genotyping.
Reliable SNP genotyping. Accurate genotyping.
Accurate genotyping.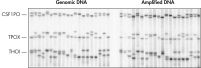 Comparable NGS (next-generation sequencing) results obtained using purified gDNA or REPLI-g amplified DNA.
Comparable NGS (next-generation sequencing) results obtained using purified gDNA or REPLI-g amplified DNA. Schematic representation of REPLI-g amplification.
Schematic representation of REPLI-g amplification. Highly representative amplification using REPLI-g technology.
Highly representative amplification using REPLI-g technology. Consistent and accurate whole genome amplification.
Consistent and accurate whole genome amplification.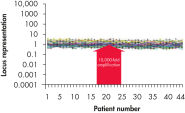
Principle
Amplification principle
Sufficient quantity of genomic DNA for genomic analysis is often lacking, restricting the analyses that can be performed. Whole genome amplification (WGA) overcomes this limitation by amplification of the entire genome within a sample, providing sufficient quantities to perform all analyses on the same DNA sample. REPLI-g technology delivers highly uniform DNA amplification across the entire genome with minimal sequence bias.
The method is based on multiple displacement amplification (MDA) technology, which uses isothermal genome amplification with a uniquely processive Phi 29 polymerase that is capable of replicating up to 100 kb without dissociating from the genomic DNA template (see figure “ Multiple displacement amplification [MDA] technology”). Phi 29 polymerase is used in the presence of exonuclease-resistant primers to achieve high yields of DNA product and has a 3'→5' exonuclease proofreading activity with 1000-fold higher fidelity than Taq polymerase to maintain high fidelity during replication. Supported by the unique, optimized REPLI-g buffer system, Phi 29 polymerase easily solves secondary structures such as hairpin loops, thereby preventing slipping, stoppage and dissociation of the polymerase during amplification. This enables the generation of DNA fragments up to 100 kb without sequence bias (see figure " Unbiased amplification with Phi 29 polymerase").
Alkaline denaturation of DNA
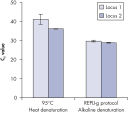
Genomic DNA must be denatured before use in enzymatic amplification procedures, which is often accomplished using harsh methods such as incubation at elevated temperatures (heat incubation) or increased pH (chemical alkaline incubation). REPLI-g Kits use gentle alkaline incubation, allowing uniform DNA denaturation with very low DNA fragmentation or generation of abasic sites. This results in amplified DNA with very high integrity, and maximizes the length of amplified fragments so that genomic loci and sequences are uniformly represented. With REPLI-g Kits, reliable results without false positive or negative data are ensured in subsequent downstream applications, unlike with other WGA technologies that use heat-induced denaturation that can damage template DNA, leading to biased and underrepresented loci (see figure " Effect of heat and alkaline denaturation on loci representation").
See figures
Procedure
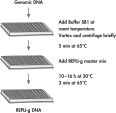
REPLI-g Mini and Midi Kits use a simple and reliable method to achieve accurate genome amplification from small quantities of isolated target genomic DNA, or directly from whole blood, dried blood cards, buffy coat and tissue culture cells (see figure " REPLI-g Mini and Midi procedure"). The addition of lysis buffer, which both lysis the sample material and denatures the DNA, is followed by a short minute incubation (see figure " REPLI-g Mini and Midi procedure"). After neutralization, master mix (including REPLI-g Mini DNA Polymerase) is added and the isothermal amplification reaction proceeds overnight at 30°C. REPLI-g amplified DNA can be stored long-term at –20°C with no negative effects (see figure " Consistent long-term stability").
Amplification of genomic DNA using the REPLI-g UltraFast Mini Kit involves 3 basic steps (see figure " Purified genomic DNA procedure"). First, the sample (10 ng purified genomic DNA, 0.5 µl whole blood, or 300 tissue culture cells) undergoes gentle alkaline denaturation, avoiding fragmentation and damage of template DNA. Next the sample is neutralized, and finally incubated with REPLI-g UltraFast DNA Polymerase at 30°C. The amplified DNA is ready to use after 60 minutes without further purification.
The very fast, simple and accurate REPLI-g Screening method is capable of parallel amplification of several genomic samples, as described in the REPLI-g Screening Handbook. Sample material is lysed and the DNA is denatured by incubating with Buffer SB1 at 65°C. Denaturation is stopped by cooling to room temperature. Buffer SB2 and DNA polymerase are added, and the isothermal amplification proceeds for at least 10 hours or overnight at 30°C.
The entire reaction setup is performed at room temperature, making it easy to use with liquid-handling instrumentation. Reaction setup for 96 samples takes only 20 minutes, saving time to concentrate on other important tasks. The REPLI-g Screening Kit combines a number of features that make it highly suitable for use in manual or automated high-throughput mutation screening, including high volume pipetting steps for increased accuracy, compatibility with 96-well microplates, and a streamlined protocol for fast and easy setup (see flowchart " REPLI-g screening procedure").
Select the REPLI-g Kit most suited to your specific requirements from our complete range of dedicated REPLI-g products (see table “Specifications for the wide range of REPLI-g Kits”).
| REPLI-g Single Cell | REPLI-g Mini | REPLI-g UltraFast Mini | REPLI-g Midi | REPLI-g Screening | REPLI-g FFPE | REPLI-g Mitochondrial DNA | |
|---|---|---|---|---|---|---|---|
| Starting material | Single cells, gDNA | Purifed gDNA, blood, cells | Purifed gDNA, blood, cells | FFPE tissue, purified gDNA from FFPE tissue | Purified gDNA | ||
| Input amount | Single cells, 2–1000 cells, tissue, purified gDNA (1–10 ng) | >10 ng gDNA, 0.5 µl blood or cells (>600 cells/µl) | >10 ng gDNA, 0.5 µl blood or cells (>600 cells/µl) | Section (1 cm diamter, 10–40 µm thick); >100 ng gDNA | >1 ng purified gDNA | ||
| Yield (µg/reaction) | 40 | 10 | 7–10 | 40 | 8 | Standard yield: ≤10; High yield: ≤40 | 3–5 |
| Reaction time | 8–16 h | 10–16 h | 1.5 h | 8–16 h | 12–16 h | Standard yield: 4 h; High yield: 10 h | 8 h |
| Hands-on time | 15 min | 15 min | 15 min | 15 min | 15 min | 40 min | 15 min |
| Format | Tube | Tube | Tube | Tube | Tube | Tube | Tube |
See figures
Applications
- REPLI-g amplified genomic can be used in a variety of downstream applications, including:
- SNP genotyping with TaqMan® primer/probe sets
- qPCR- and PCR-based mutation detection
- Next-generation sequencing
- STR/microsatellite analysis
- Sanger sequencing
- RFLP and Southern blot analysis
- Array technologies, such as comparative genomic hybridization
Comparison of REPLI-g Kits
| Features | REPLI-g UltraFast Mini Kit | REPLI-g Mini Kit | REPLI-g Midi Kit | REPLI-g Screening Kit |
| Amplification | Whole genomic DNA | Whole genomic DNA | Whole genomic DNA | Whole genomic DNA |
| Applications | Genotyping, microarray, PCR, real-time PCR, NGS | Genotyping, hybridization, RFLP, NGS | Genotyping, hybridization, RFLP, NGS | Genotyping, sequencing |
| Denaturation step | Alkaline | Alkaline | Alkaline | Heat |
| Handling time for 96 samples | – | – | – | 20 minutes |
| Maximum input volume | 10 ng DNA, 0.5 µl whole blood, ~300 cells/µl | >10 ng DNA, 0.1– 0.5 µl whole blood, >600 cells/µl | >10 ng DNA, 0.1–0.5 µl whole blood, >600 cells/µl | 10 ng DNA, 0.5 µl whole blood, >600 cells/µl |
| Minimal pipetting volume needed | 1 µl | 0.5 µl | 0.5 µl | 1 µl |
| Quality assessment | No | No | No | No |
| Reaction time | 60–90 minutes | 10–16 hours | 8–16 hours | 10–16 hours |
| Reaction volume | 20 µl | 50 µl | 50 µl | 40 µl |
| Samples per run (throughput) | Medium | Medium | Medium | High (1–96 samples) |
| Starting amount of DNA | >10 ng purified genomic DNA | >10 ng purified genomic DNA | >10 ng purified genomic DNA | >10 ng purified genomic DNA |
| Starting material | Genomic DNA, blood, cells | Genomic DNA, blood, cells, tissue | Genomic DNA, blood, cells, tissue | Genomic DNA, blood, cells |
| Technology | Multiple Displacement Amplification (MDA) | Multiple Displacement Amplification (MDA) | Multiple Displacement Amplification (MDA) | Multiple Displacement Amplification (MDA) |
| Yield | 7–10 µg | 10 µg | 40 µg | ~8 µg |
Supporting data and figures
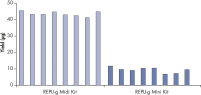
Consistent DNA yields using any sample type.
Various starting materials, including genomic DNA, and heparin- and EDTA-preserved whole blood, were amplified using REPLI-g Midi and Mini Kits. Typical yields of 40 µg (Midi Kit) and 8–10 µg (Mini Kit) were obtained.