Welcome to QIAGEN!
You can accept or revoke the cookies used on this website at any time with the selection below or by adjusting your cookie settings. For complete details about our cookies, see our Cookie Policy.
用于分析特定位点的拷贝数变化和改变
目录编号 / ID. 337812
qBiomarker Copy Number PCR Assays可对单个基因或目的区域(GOI或ROI)的拷贝数变化进行特定、准确、可重复、易解读的分析。每次分析都经实验验证,可即用于微阵列研究、特定靶标筛选和相关研究。
qBiomarker Copy Number PCR Assays非常适合准确检测新鲜、冷冻或福尔马林固定石蜡包埋(FFPE)样本的单个位点的拷贝数改变或变化。
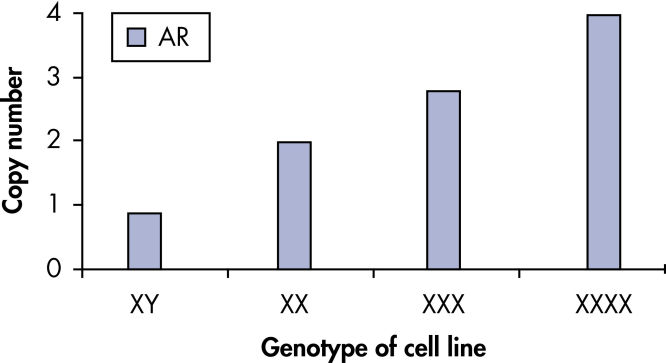
qBiomarker Copy Number PCR Assays accurately identify aneuploidy.