✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
EpiTect PCR Control DNA Set (100)
Cat. No. / ID: 59695
✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 使用便利、经严格质量控制的即用型DNA溶液
- 亚硫酸氢盐转化的DNA,用于对照实验
- 适用于各种甲基化分析
产品详情
EpiTect Control DNAs是即用型、完全甲基化或非甲基化经亚硫酸氢盐转化的DNA,以及未处理的非甲基化基因组DNA,为甲基化分析设立标准可靠的对照反应。甲基化或未甲基化的亚硫酸氢盐转化DNA储存在EB Buffer(10 mM Tris·Cl)中,是浓度为10 ng/μl的即用型溶液。未转化的非甲基化人类对照DNA也储存在EB Buffer(10 mM Tris·Cl)中,是浓度为50 ng/μl的即用型溶液。
绩效
查看图表
原理
查看图表
程序
表观遗传学中标准的工作流程
表观遗传学信息不仅在生物和医学研究领域,尤其肿瘤学研究中是非常重要的,而且对干细胞研究和生物学的发展也至关重要。由于缺乏从有限样本中获得可重复数据的标准方法,分析DNA甲基化变化仍是一个难题。配合其新引进的EpiTect溶液,QIAGEN提供标准的分析前和分析溶液,用于从DNA样本收集、稳定和纯化到亚硫酸氢盐转化和real-time或终点PCR甲基化分析或测序(参见" Standardized workflows in epigenetics")。
查看图表
应用
EpiTect Control DNAs适用于各种甲基化分析的对照反应,包括:
- 评估MSP引物的特异性
- 作为HRM和MethyLight PCR的定量标准
- 评估MethyLight PCR引物和探针的特异性
- 确定亚硫酸氢盐转化反应的效率
辅助数据和图表
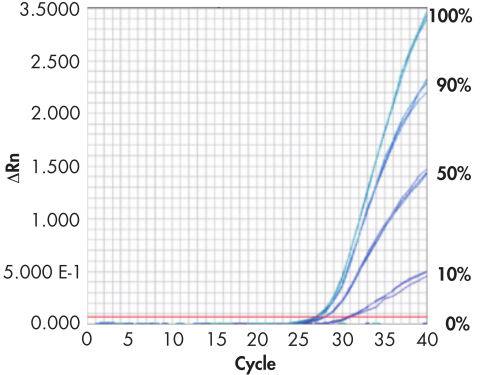
Real-time PCR standards.
EpiTect Methylated Control DNA and EpiTect Unmethylated Control DNA (both pre-bisulfite converted) were mixed to give 100%, 90%, 50%, 10%, and 0% methylated DNA. EpiTect MethyLight Assays for the human PITX2 gene were run in triplicate, using 10 ng of each DNA sample. The results show that EpiTect Control DNA can be used as a methylation standard for the quantification of unknown DNA samples.
Specifications
| Features | Specifications |
|---|---|
| ApplicationsZH | End point Methylation Specific PCR (MSP), real-time methylation specific PCR |
| Number of reactions | for 1000 control PCRs |
| Concentration | 10 ng/µl (1 µg in total) excepted the unmethylated control DNA: 50 ng/µl (10 µg in total) |