QuantiFast Multiplex RT-PCR Kits
使用序列特异性探针进行快速的多重一步法RT-PCR,适用于基因表达分析
使用序列特异性探针进行快速的多重一步法RT-PCR,适用于基因表达分析

Cat. No. / ID: 204854
应用QuantiFast Multiplex RT-PCR Kits可在单管内通过多重一步法real-time RT-PCR对多至4个RNA进行快速、可靠的定量检测。特制的逆转录酶混合液可快速、高效合成cDNA。Q-Bond和优化的预混液促进快速多重real-time RT-PCR的进行,适用于快速或标准PCR仪。即用型预混液中的热启动酶和独特的PCR缓冲液可确保在所有real-time PCR仪上进行灵敏的qPCR,无需优化。有两种规格的试剂盒:QuantiFast Multiplex RT-PCR Kit适用于需要ROX染料进行荧光信号校准的PCR仪,QuantiFast Multiplex RT-PCR +R Kit适用于其他所有PCR仪。为便于使用,预混液可保存在2–8°C。
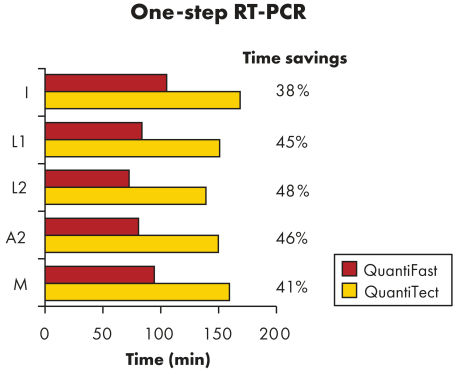
QuantiFast Multiplex RT-PCR Kits可节省高达50%的RT-PCR运行时间,让您更快获得结果(参见"Significantly reduced RT-PCR times")。您可极大地提高样本通量或与其它使用者有效地共享PCR仪。在同一个反应中同时扩增对照基因和靶基因,而非分开反应,减少了操作误差,提高了基因定量检测的可靠性(参见"Reliable relative quantification")。QuantiFast Multiplex RT-PCR Kits提供的特制预混液可快速构建多重反应,通常初次使用即可得到成功的结果,且多重PCR反应结果可与单重PCR反应结果相媲美(参见"Comparable results in triplex and singleplex RT-PCR")。
QuantiFast Multiplex RT-PCR Kits可清楚区分模板量的细微差别。即使模板量仅有两倍的差别,该试剂盒也能精确定量丰度差别较大的2个靶基因。该试剂盒可对多达4个靶基因进行快速多重real-time RT-PCR检测,且不会影响PCR结果(参见"Uncompromised sensitivity in 4-plex RT-PCR")。
QuantiFast Multiplex RT-PCR Kits在标准和快速PCR仪上快速、灵敏的进行检测,无需优化(参见"QIAGEN multiplex kits")。特制的快速PCR缓冲液中含有新型成分Q-Bond,可大大缩短变性、退火和延伸所需时间(参见"Fast primer annealing")。
在同一个反应中同时扩增对照基因和靶基因,而非分开反应,减少了操作误差,提高了基因定量检测的可靠性。QuantiFast Multiplex RT-PCR Buffer含有平衡的K+和NH4+配比,促进了引物的特异性退火,同时独特的MP因子可稳定特异性结合的引物(参见"Unique PCR buffer")。此外,经优化的逆转录酶混合液确保cDNA合成在20分钟内完成。HotStarTaq Plus DNA Polymerase要求严谨的热启动,可阻止非特异性产物的形成。
| 成分 | 特点 | 优势 |
|---|---|---|
| HotStarTaq Plus DNA Polymerase | 95ºC加热5分钟活化 | 在室温进行qPCR反应体系构建 |
| QuantiFast Multiplex RT-PCR Buffer | 平衡的NH4+和K+离子配比 | 引物探针的特异性结合确保获得可靠结果 |
| 合成的MP因子 | 在单管中对至多4个基因进行多重分析 | |
| 独特的Q-Bond添加剂 | PCR运行时间缩短,更快获得结果,一天内可完成更多PCR反应 | |
| ROX染料† | 对Applied Biosystems和Agilent PCR仪进行荧光信号的校准 | 在需要ROX染料的PCR仪上进行准确定量,不影响PCR反应结果 |
| QuantiFast RT Mix | 特制的逆转录酶混合液,对RNA具有高度亲和性 | RNA的逆转录可在20分钟内完成,包括复杂的二级结构 |
QuantiFast Multiplex RT-PCR Kits提供即用型的预混液,无需优化反应及循环条件。只需在预混液中加入模板RNA和引物-探针对,即可按照操作手册中的实验方案,在任何real-time PCR仪上进行快速、可靠的检测。试剂盒提供两种形式:预混液中含有或不含荧光校正用的ROX染料,因此适用于任何real-time PCR仪(参见下表)。由于ROX浓度经优化,即使是低拷贝数也可通过数据自动分析进行检测。
| ROX染料 | 试剂盒 | 适用的PCR仪 |
|---|---|---|
| 混合在预混液中 | QuantiFast Multiplex RT-PCR Kit | 除Applied Biosystems 7500外的所有Applied Biosystems仪器 |
| 单独装在管中 | QuantiFast Multiplex RT-PCR +R Kit | Applied Biosystems 7500、 Bio-Rad、Cepheid、Eppendorf、QIAGEN、Roche、Agilent和其他供应商的PCR仪 |
QuantiFast Multiplex RT-PCR Kits可用于在各种real-time PCR仪上进行基因表达分析,包括Applied Biosystems、Bio-Rad、Cepheid、Eppendorf、Roche和Agilent的PCR仪。Rotor-Gene Q实时荧光定量PCR分析仪或其他Rotor-Gene PCR仪上使用时,我们推荐使用专为快速PCR研发的Rotor-Gene Multiplex RT-PCR Kit。
| Features | Specifications |
|---|---|
| ApplicationsZH | Real-time quantification of RNA targets in a multiplex format |
| Reaction type | Real-time one-step RT-PCR |
| Single or multiplex | Multiplex |
| SYBR Green I or sequence-specific probes | Sequence-specific probes |
| Real-time or endpoint | Real-time |
| Thermal cycler | Real-time cyclers dedicated for multiplex PCR (e.g., most Applied Biosystems real-time PCR cyclers, Roche LightCycler 480, and Bio-Rad iCycler iQ) |
| Sample/target type | RNA |
| With or without ROX | Available with ROX in master mix or with ROX as separate vial |